Search Count: 24
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PROTEIN FIBRIL |
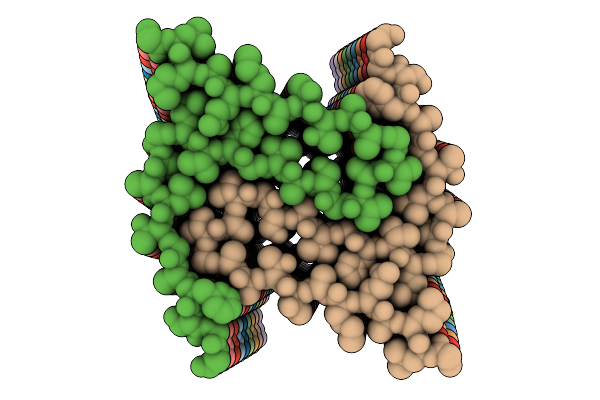 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PROTEIN FIBRIL |
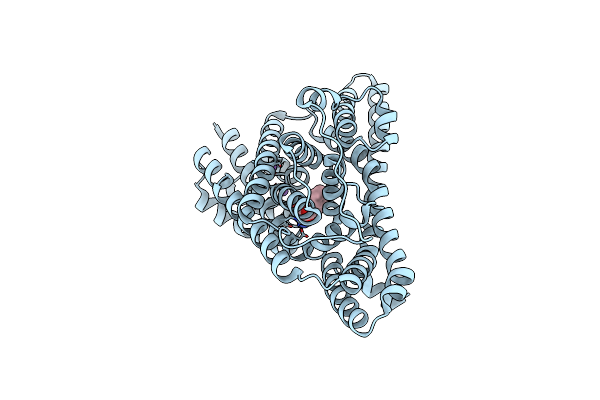 |
Organism: Pyrococcus horikoshii
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: TB1, NA |
 |
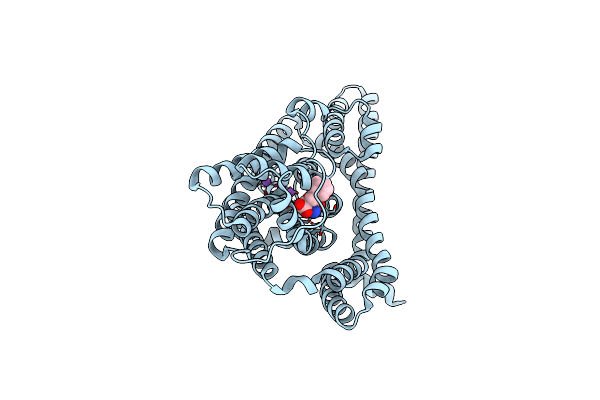
Organism: Pyrococcus horikoshii
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: TRANSPORT PROTEIN Ligands: ASP, NA |
 |
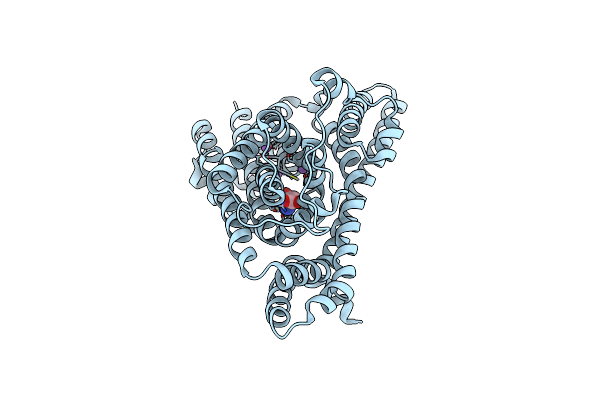
Organism: Pyrococcus horikoshii
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: TRANSPORT PROTEIN Ligands: TB1, NA |
 |
Organism: Pyrococcus horikoshii
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: TRANSPORT PROTEIN Ligands: ASP, NA |
 |
Organism: Pyrococcus horikoshii
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: TRANSPORT PROTEIN Ligands: ASP, NA |
 |
Organism: Pyrococcus horikoshii
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: TRANSPORT PROTEIN Ligands: ASP, NA |
 |
Organism: Pyrococcus horikoshii
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: TRANSPORT PROTEIN Ligands: ASP, NA |
 |
Organism: Pyrococcus horikoshii
Method: ELECTRON MICROSCOPY Release Date: 2020-06-03 Classification: TRANSPORT PROTEIN Ligands: ASP, 6OU |
 |
Gltph In Complex With L-Aspartate And Sodium Ions In Intermediate Outward-Facing State
Organism: Pyrococcus horikoshii
Method: ELECTRON MICROSCOPY Release Date: 2020-04-22 Classification: TRANSPORT PROTEIN Ligands: ASP, 6OU |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-01-31 Classification: LIPID TRANSPORT |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-01-17 Classification: LIPID TRANSPORT Ligands: HC3 |
 |
Organism: Streptomyces lividans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-07-13 Classification: METAL TRANSPORT Ligands: K |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-07-22 Classification: LIPID BINDING PROTEIN |

