Search Count: 20
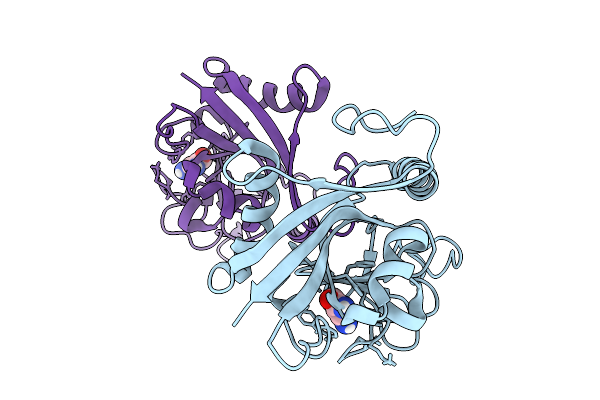 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: 3AB |
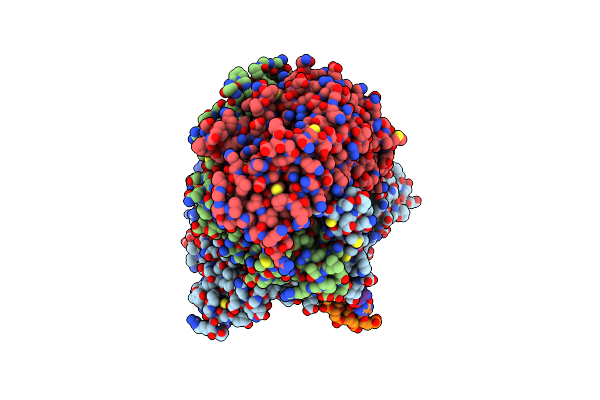 |
Structure Of The G848S Mutant Of Human Mitochondrial Dna Polymerase Gamma In Complex With Pzl-A
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSFERASE/DNA Ligands: CA, DCP, A1IK1 |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSFERASE/DNA Ligands: CA, DCP |
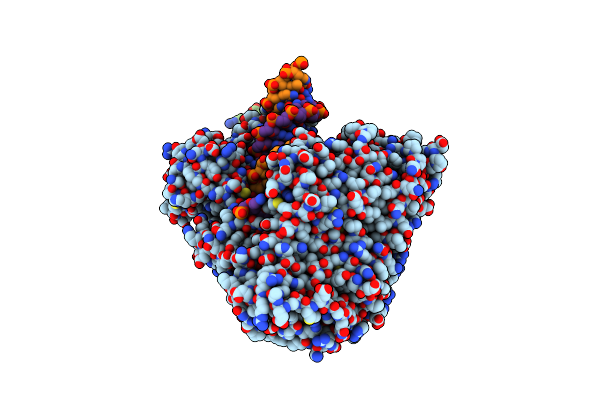 |
Structure Of The A467T Mutant Of Human Mitochondrial Dna Polymerase Gamma In Complex With Pzl-A
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSFERASE/DNA Ligands: CA, DCP, A1IK1 |
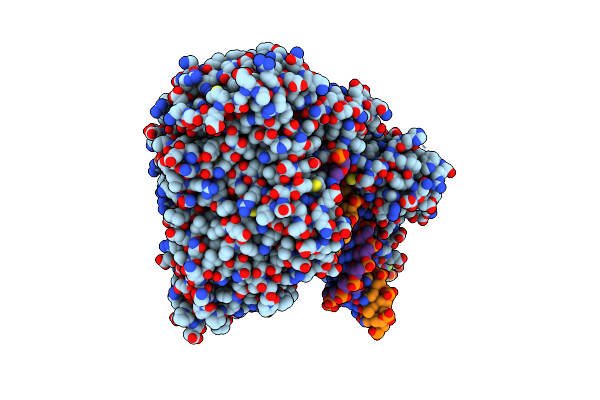 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSFERASE/DNA Ligands: CA, DCP |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSFERASE/DNA Ligands: CA, DCP |
 |
Cryo-Em Structure Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri In Its Tetrameric State Produced In The Presence Of Datp And Ctp
Organism: Segatella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: BIOSYNTHETIC PROTEIN Ligands: DTP, MG |
 |
Cryo-Em Structure Of The Dimeric Form Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri Produced In The Presence Of Datp And Ctp
Organism: Segatella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: BIOSYNTHETIC PROTEIN Ligands: DTP, MG |
 |
Cryo-Em Structure Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri In Its Dimeric, Atp/Dttp/Gtp-Bound State
Organism: Segatella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: BIOSYNTHETIC PROTEIN Ligands: TTP, MG, GTP, ZN |
 |
Cryo-Em Structure Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri In Its Dimeric, Dgtp/Atp-Bound State
Organism: Segatella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: BIOSYNTHETIC PROTEIN Ligands: DGT, MG, ATP, ZN |
 |
Targeting Toll-Like Receptor-Driven Systemic Inflammation By Engineering An Innate Structural Fold Into Drugs
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2023-09-06 Classification: DE NOVO PROTEIN |
 |
Cryo-Em Structure Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri In Its Dimeric, Atp/Ctp-Bound State
Organism: Prevotella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: BIOSYNTHETIC PROTEIN Ligands: ATP, CTP, ZN |
 |
Cryo-Em Structure Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri In Its Dimeric, Datp-Bound State
Organism: Segatella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: BIOSYNTHETIC PROTEIN Ligands: DTP, MG |
 |
Cryo-Em Structure Of The Anaerobic Ribonucleotide Reductase From Prevotella Copri In Its Tetrameric, Datp-Bound State
Organism: Segatella copri
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: OXIDOREDUCTASE Ligands: DTP, MG |
 |
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: VIRAL PROTEIN |
 |
Organism: Bacillus subtilis (strain 168), Geobacillus kaustophilus (strain hta426)
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2019-04-24 Classification: PROTEIN BINDING Ligands: MG |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168, Geobacillus kaustophilus hta426
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-04-24 Classification: PROTEIN BINDING Ligands: CL |

