Search Count: 108
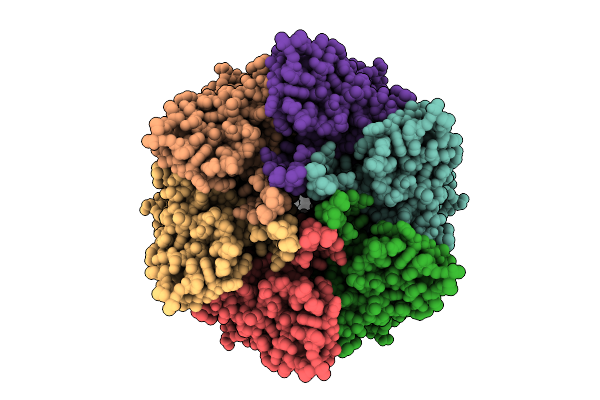 |
Intermembrane Lipid Transport Complex Letab From Escherichia Coli (Composite Model Corresponding To Map 2)
Organism: Escherichia coli str. k-12 substr. mg1655
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: LIPID TRANSPORT Ligands: ZN, PEF |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: DE NOVO PROTEIN |
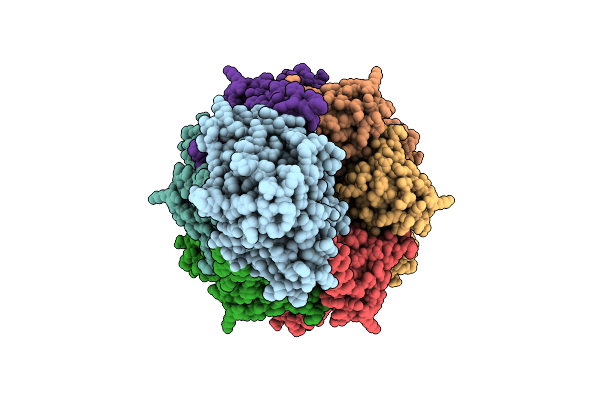 |
Intermembrane Lipid Transport Complex Letab From Escherichia Coli (Crosslinked, Composite Model Corresponding To Map 1)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: LIPID TRANSPORT Ligands: ZN |
 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Mycobacterium tuberculosis, Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: DE NOVO PROTEIN |
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-10-25 Classification: UNKNOWN FUNCTION Ligands: GOL, EDO |
 |
Crystal Structure Of The N-Terminal Domain Of The Cryptic Surface Protein (Cd630_25440) From Clostridium Difficile.
Organism: Clostridioides difficile 630
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-05-10 Classification: UNKNOWN FUNCTION Ligands: CL, EDO, GOL |
 |
Crystal Structure Of Designed Helical Repeat Protein Rpb_Plp3_R6 Bound To Plpx6 Peptide
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2023-03-29 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Helical Repeat Protein Rpb_Pew3_R4 Bound To Pawx4 Peptide
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-03-22 Classification: DE NOVO PROTEIN |
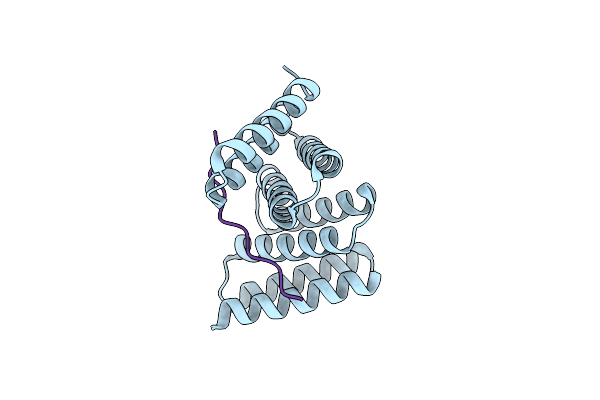 |
Crystal Structure Of Designed Helical Repeat Protein Rpb_Lrp2_R4 Bound To Lrpx4 Peptide
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2023-03-22 Classification: DE NOVO PROTEIN |
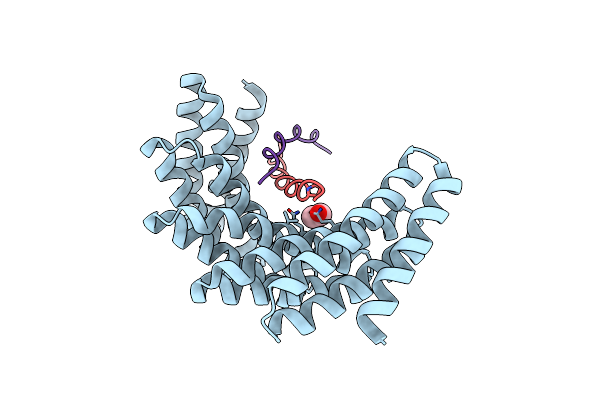 |
Crystal Structure Of Designed Helical Repeat Protein Rpb_Plp1_R6 Bound To Plpx6 Peptide
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-03-22 Classification: DE NOVO PROTEIN Ligands: EDO |
 |
Crystal Structure Of Designed Helical Repeat Protein Rpb_Plp1_R6 In Alternative Conformation 1 (With Peptide)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-03-22 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Helical Repeat Protein Rpb_Plp1_R6 In Alternative Conformation 2
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-03-22 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Helical Repeat Protein Rpb_Lrp2_R4 (Proteolysis Fragment?), Forming Pseudopolymeric Filaments
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-03-22 Classification: DE NOVO PROTEIN Ligands: EDO, PO4 |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: MEMBRANE PROTEIN Ligands: UNL |
 |
Structure Of Mce1 Transporter From Mycobacterium Smegmatis In The Absence Of Lucb (Map2)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: MEMBRANE PROTEIN Ligands: UNL |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: MEMBRANE PROTEIN Ligands: UNL |

