Search Count: 22
 |
Cryo-Em Structure Of Tmprss2 In Complex With Fab Fragments Of 752 Mab And 2228 Mab
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
 |
Organism: Burkholderia
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-05-14 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE |
 |
Organism: Burkholderiales bacterium
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, SCN |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab354
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab159
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN Ligands: NAG |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab188
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN Ligands: NAG |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab326
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With An Fv-Clasp Form Of A Human Neutralizing Antibody Ab496
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab445
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Human Dnmt1 (Aa:351-1616) In Complex With Ubiquitinated H3 And Hemimethylated Dna Analog (Cxxc-Ordered Form)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: TRANSFERASE/DNA Ligands: ZN, SAH |
 |
Cryo-Em Structure Of Human Dnmt1 (Aa:351-1616) In Complex With Ubiquitinated H3 And Hemimethylated Dna Analog (Cxxc-Disordered Form)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: TRANSFERASE/DNA Ligands: ZN |
 |
X-Ray Crystal Structure Of Cyclic-Pip And Dna Complex In A Reverse Binding Orientation
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-06-24 Classification: DNA Ligands: S7E, MG, NA, EDO |
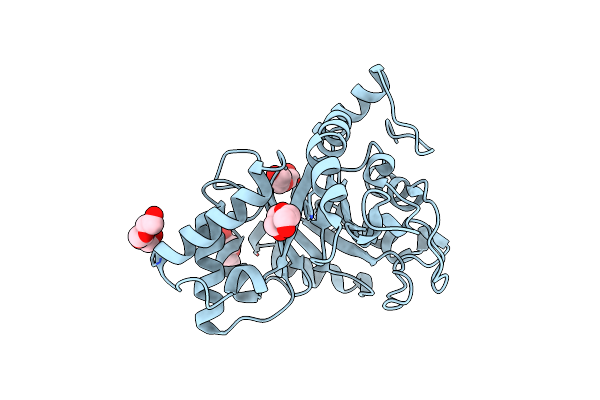 |
Crystal Structure Of Endo-Beta-N-Acetylglucosaminidase From Cordyceps Militaris In Apo Form
Organism: Cordyceps militaris cm01
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: PGE, PEG |
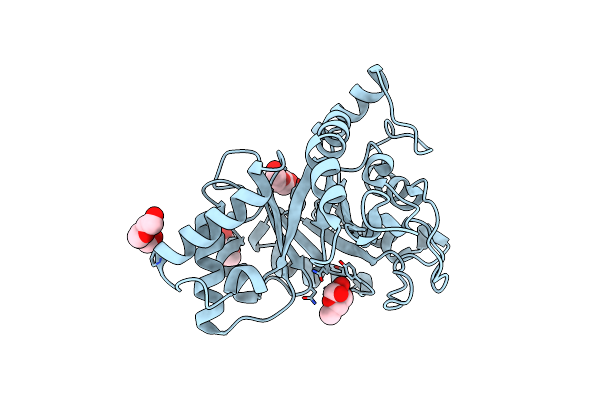 |
Crystal Structure Of Endo-Beta-N-Acetylglucosaminidase From Cordyceps Militaris In Complex With L-Fucose
Organism: Cordyceps militaris cm01
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: FUC, PGE, PEG |
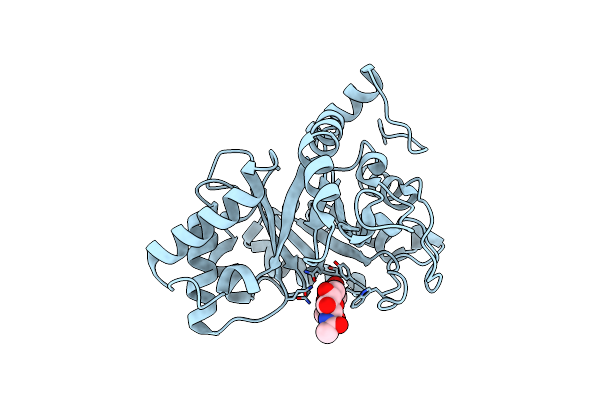 |
Crystal Structure Of Endo-Beta-N-Acetylglucosaminidase From Cordyceps Militaris D154N/E156Q Mutant In Complex With Fucosyl-N-Acetylglucosamine
Organism: Cordyceps militaris cm01
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-10-02 Classification: HYDROLASE |
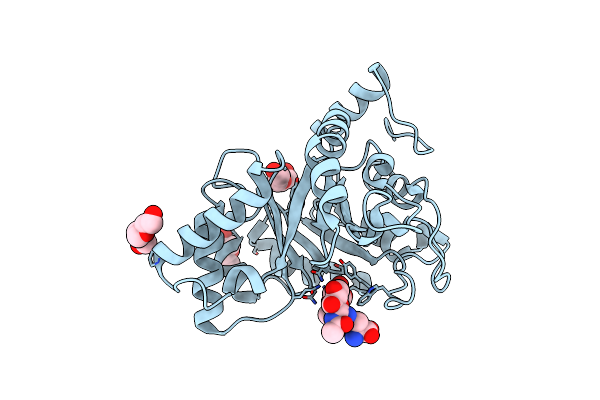 |
Crystal Structure Of Endo-Beta-N-Acetylglucosaminidase From Cordyceps Militaris D154N/E156Q Mutant In Complex With Fucosyl-N-Acetylglucosamine-Asn
Organism: Cordyceps militaris cm01
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: PGE, PEG, ASN |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:0.94 Å Release Date: 2019-04-17 Classification: FLUORESCENT PROTEIN Ligands: CL |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2019-04-17 Classification: FLUORESCENT PROTEIN |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:0.78 Å Release Date: 2019-04-17 Classification: FLUORESCENT PROTEIN Ligands: MG |

