Search Count: 9
 |
Crystal Structure Of The S. Cerevisiae Eif2Beta N-Terminal Tail Bound To The C-Terminal Domain Of Eif5
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSLATION |
 |
Organism: Synechococcus elongatus pcc 6301, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-12-14 Classification: LYASE/DNA Ligands: MG, FAD, HDF |
 |
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-04-13 Classification: LYASE Ligands: PO4, FAD |
 |
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-04-13 Classification: LYASE Ligands: PO4, FAD |
 |
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-04-13 Classification: LYASE Ligands: PO4, FAD |
 |
Data4:Photoreduced Dna Photolyase / Received X-Rays Dose 1.2 Exp15 Photons/Mm2
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-04-13 Classification: LYASE Ligands: PO4, FAD |
 |
Data6:Photoreduced Dna Pholyase / Received X-Rays Dose 4.8 Exp15 Photons/Mm2
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-04-13 Classification: LYASE Ligands: PO4, FAD |
 |
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1998-01-14 Classification: DNA REPAIR Ligands: FAD, HDF |
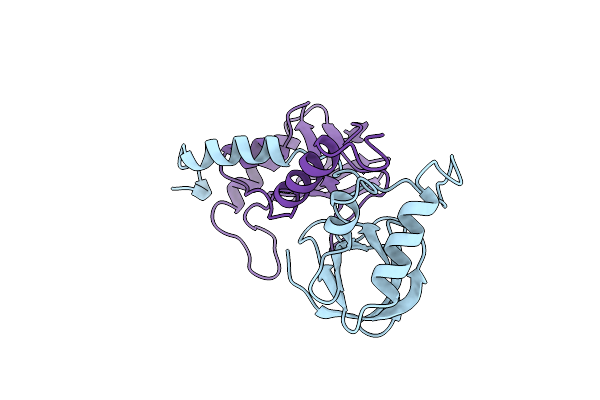 |
Staphylococcal Nuclease Dimer Containing A Deletion Of Residues 114-119 Complexed With Calcium Chloride And The Competitive Inhibitor Deoxythymidine-3',5'-Diphosphate
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 1997-04-21 Classification: HYDROLASE |

