Search Count: 33
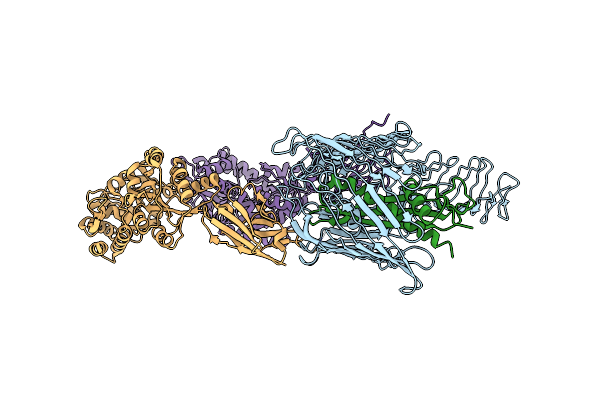 |
Cryo-Em Structure Of N. Gonorhoeae Lptde In Complex With Promacrobodies (Mbps Have Not Been Built De Novo)
Organism: Neisseria gonorrhoeae, Synthetic construct, Methanosarcina mazei
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: TRANSPORT PROTEIN |
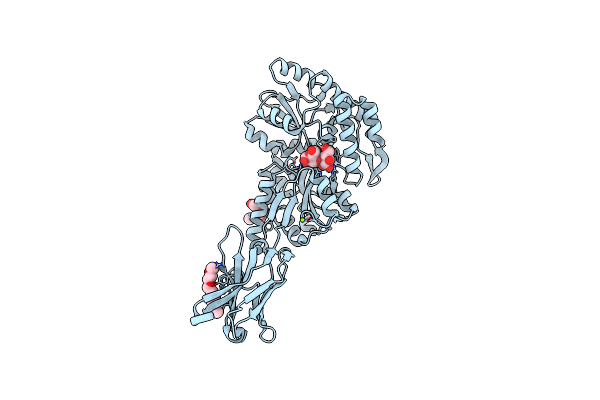 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-05-04 Classification: IMMUNE SYSTEM Ligands: P6G, MG |
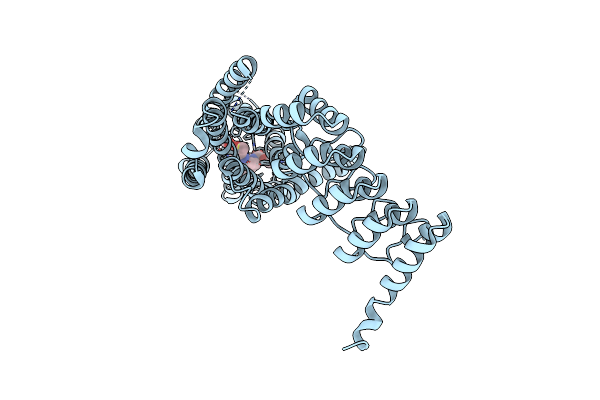 |
Crystal Structure Of The Human Alpha1B Adrenergic Receptor In Complex With Inverse Agonist (+)-Cyclazosin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2022-01-12 Classification: MEMBRANE PROTEIN Ligands: T0B |
 |
Sars-Cov-2 Spike Protein In Complex With Sybody#15 And Sybody#68 In A 3Up Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: VIRAL PROTEIN Ligands: NAG |
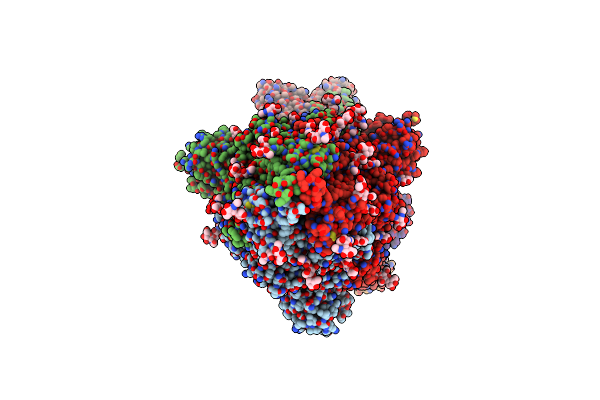 |
Sars-Cov-2 Spike Protein In Complex With Sybody#15 And Sybody#68 In A 1Up/1Up-Out/1Down Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: VIRAL PROTEIN Ligands: NAG |
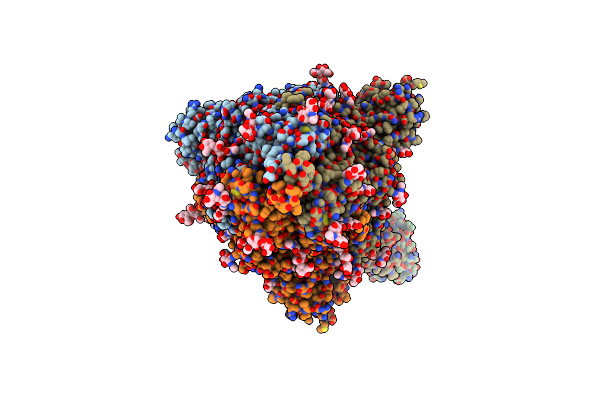 |
Sars-Cov-2 Spike Protein In Complex With Sybodyb#15 In A 1Up/1Up-Out/1Down Conformation.
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: VIRAL PROTEIN Ligands: NAG |
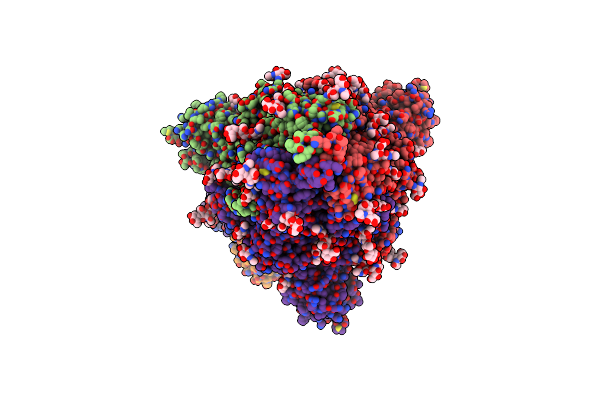 |
Sars-Cov-2 Spike Protein In Complex With Sybody#68 In A 2Up/1Flexible Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: VIRAL PROTEIN Ligands: NAG |
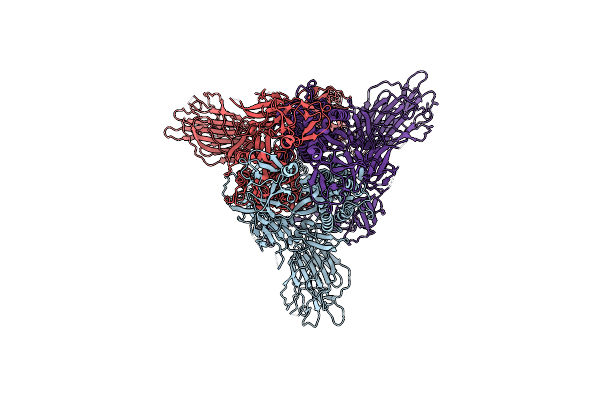 |
Sars-Cov-2 Spike Protein In Complex With Sybody No68 In A 1Up/2Down Conformation
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of The Neurotensin Receptor 1 In Complex With The Peptide Full Agonist Nts8-13
Organism: Rattus norvegicus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2021-02-10 Classification: MEMBRANE PROTEIN Ligands: BNG |
 |
Crystal Structure Of The Neurotensin Receptor 1 In Complex With The Small-Molecule Inverse Agonist Sr142948A
Organism: Rattus norvegicus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2021-02-10 Classification: MEMBRANE PROTEIN Ligands: Q6N |
 |
Crystal Structure Of The Neurotensin Receptor 1 (Ntsr1-H4Bmx) In Complex With The Small Molecule Inverse Agonist Sr48692
Organism: Rattus norvegicus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2021-02-10 Classification: MEMBRANE PROTEIN Ligands: Q6Q |
 |
Crystal Structure Of The Neurotensin Receptor 1 (Ntsr1-H4Bmx) In Complex With Nts8-13
Organism: Rattus norvegicus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-02-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Rattus norvegicus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2021-02-10 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The Neurotensin Receptor 1 In Complex With The Small-Molecule Full Agonist Sri-9829
Organism: Rattus norvegicus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-02-10 Classification: MEMBRANE PROTEIN Ligands: SR9 |
 |
Crystal Structure Of The Neurotensin Receptor 1 In Complex With The Small-Molecule Partial Agonist Rti-3A
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2021-02-10 Classification: MEMBRANE PROTEIN Ligands: SR5 |
 |
Crystal Structure Of The Neurotensin Receptor 1 In Complex With The Small Molecule Inverse Agonist Sr48692
Organism: Rattus norvegicus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2021-02-10 Classification: MEMBRANE PROTEIN Ligands: Q6Q |
 |
Organism: Mycolicibacterium thermoresistibile, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-04-01 Classification: MEMBRANE PROTEIN Ligands: NI, DMU |
 |
Organism: Mycolicibacterium thermoresistibile
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-01 Classification: MEMBRANE PROTEIN Ligands: FAD |
 |
Structure Of Atpgs-Bound Outward-Facing Tm287/288 In Complex With Sybody Sb_Tm35
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2019-05-29 Classification: MEMBRANE PROTEIN Ligands: AGS, MG |
 |
Structure Of Atp-Bound Outward-Facing Tm287/288 In Complex With Sybody Sb_Tm35
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2019-05-29 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |

