Search Count: 31
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2025-10-29 Classification: PROTEIN TRANSPORT |
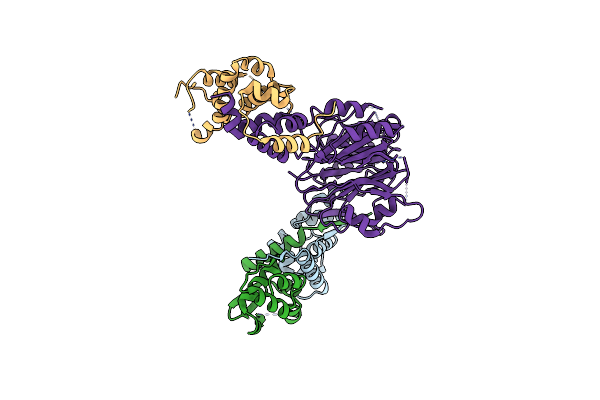 |
Cryo-Em Structure Of Skp1-Fbxo22 In Complex With A Bach1 Btb Dimer At 3.2A Resolution
|
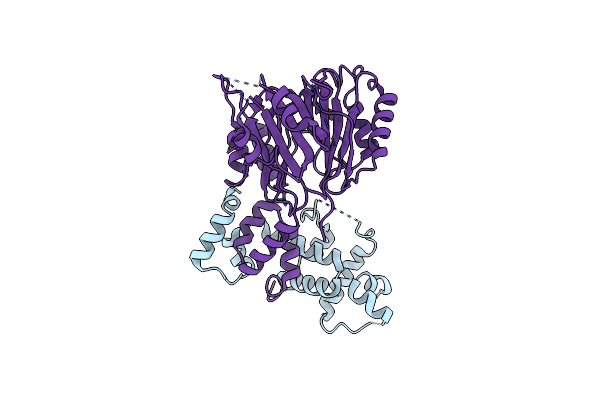 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-11 Classification: DNA BINDING PROTEIN |
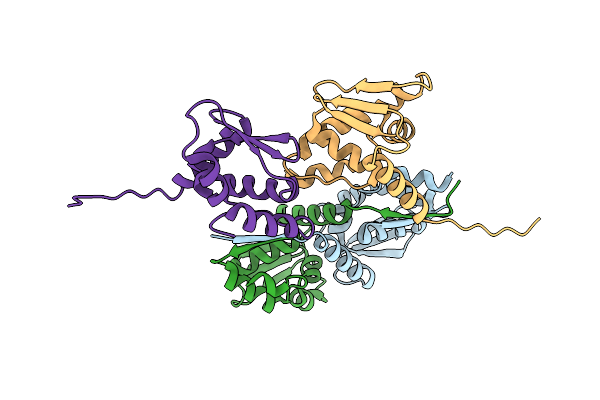 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-12-11 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-12-11 Classification: DNA BINDING PROTEIN Ligands: MRD |
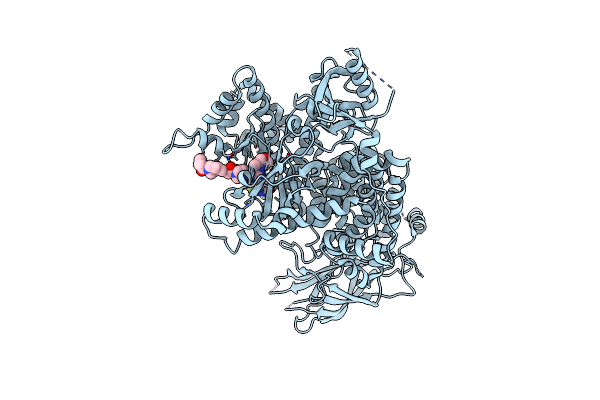 |
Structure Of P110 Alpha Bound To (S)-1-(4-((2-(4-(4-(2-Amino-4-(Difluoromethyl)Pyrimidin-5-Yl)-6-(3-Methylmorpholino)-1,3,5- Triazin-2-Yl)Piperazin-1-Yl)-2-Oxoethoxy)Methyl)Piperidin-1-Yl)Prop-2-En-1-One (Compound 9)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2024-08-28 Classification: TRANSFERASE/Inhibitor Ligands: RNX |
 |
Conformations Of Macrocyclic Peptides Sampled By Exact Noes: Models For Cell-Permeability. Nmr Structure Of Omphalotin A In Methanol / Water Indoleout Conformation.
Organism: Omphalotus olearius
Method: SOLUTION NMR Release Date: 2023-12-13 Classification: UNKNOWN FUNCTION |
 |
Conformations Of Macrocyclic Peptides Sampled By Exact Noes: Models For Cell-Permeability
Organism: Tolypocladium inflatum
Method: SOLUTION NMR Release Date: 2023-12-06 Classification: IMMUNOSUPPRESSANT |
 |
Conformations Of Macrocyclic Peptides Sampled By Exact Noes: Models For Cell-Permeability. Conformation 1 Of Omphalotin A In Apolar Solvents.
Organism: Omphalotus olearius
Method: SOLUTION NMR Release Date: 2023-12-06 Classification: UNKNOWN FUNCTION |
 |
Conformations Of Macrocyclic Peptides Sampled By Exact Noes: Models For Cell-Permeability. Nmr Structure Of Omphalotin A In Methanol / Water Indoleout Conformation.
Organism: Omphalotus olearius
Method: SOLUTION NMR Release Date: 2023-12-06 Classification: UNKNOWN FUNCTION |
 |
Organism: Omphalotus olearius
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: ZPR, DMS, BCT, EDO, GOL, PO4, NA |
 |
Organism: Omphalotus olearius
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: EDO, BCT, PEG, NA |
 |
Organism: Omphalotus olearius
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-07-06 Classification: LYASE Ligands: EDO, NA, BCT, GOL |
 |
Organism: Omphalotus olearius
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-07-06 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2016-04-13 Classification: TRANSFERASE/INHIBITOR Ligands: NVX, MPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2016-04-13 Classification: TRANSFERASE/INHIBITOR Ligands: NVV, MPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2016-04-13 Classification: TRANSFERASE Ligands: S3N, MPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2016-04-13 Classification: TRANSFERASE Ligands: 4L7, BR, MPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-04-13 Classification: TRANSFERASE Ligands: 8BQ, MPD |

