Search Count: 12
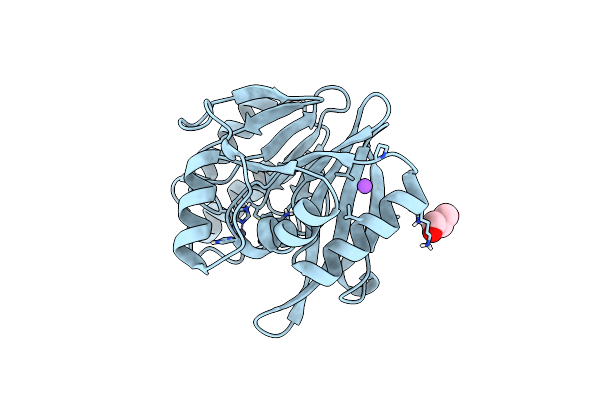 |
Native Di-Zinc Vim-26. Leu224 In Vim-26 From Klebsiella Pneumoniae Has Implications For Drug Binding.
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2015-02-04 Classification: HYDROLASE |
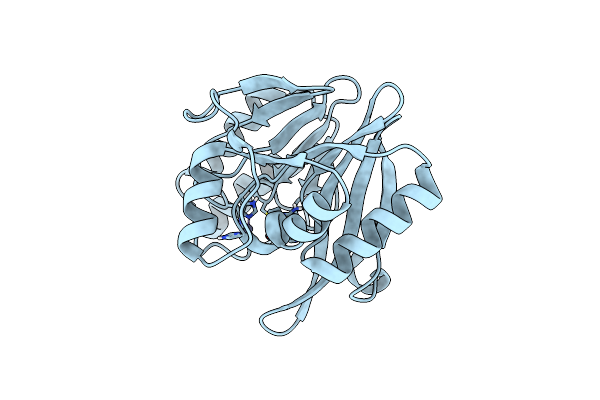 |
Penta Zn1 Coordination. Leu224 In Vim-26 From Klebsiella Pneumoniae Has Implications For Drug Binding.
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-02-04 Classification: HYDROLASE Ligands: ZN |
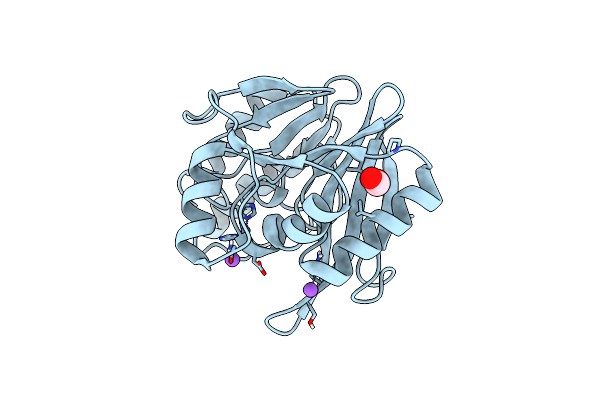 |
Mono-Zinc Vim-26. Leu224 In Vim-26 From Klebsiella Pneumoniae Has Implications For Drug Binding.
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-02-04 Classification: HYDROLASE |
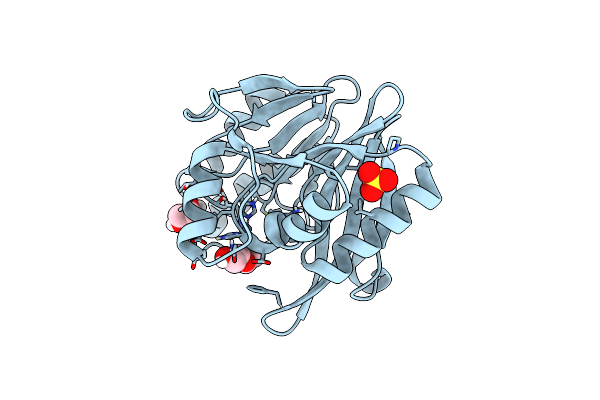 |
Vim-26-Peg. Leu224 In Vim-26 From Klebsiella Pneumoniae Has Implications For Drug Binding.
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2015-02-04 Classification: HYDROLASE Ligands: SO4, ZN, PEG, EDO |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of The Mobile Metallo-Beta-Lactamase Aim-1 From Pseudomonas Aeruginosa: Insights Into Antibiotic Binding And The Role Of Gln157
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-06-20 Classification: HYDROLASE Ligands: ZN, MG, CA |
 |
Crystal Structure Of The Mobile Metallo-Beta-Lactamase Aim-1 From Pseudomonas Aeruginosa: Insights Into Antibiotic Binding And The Role Of Gln157
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-06-20 Classification: HYDROLASE Ligands: CA, MG, ZN |
 |
Q157A Mutant. Crystal Structure Of The Mobile Metallo-Beta-Lactamase Aim-1 From Pseudomonas Aeruginosa: Insights Into Antibiotic Binding And The Role Of Gln157
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2012-06-20 Classification: HYDROLASE Ligands: ZN, CA, ACT |
 |
Q157N Mutant. Crystal Structure Of The Mobile Metallo-Beta-Lactamase Aim-1 From Pseudomonas Aeruginosa: Insights Into Antibiotic Binding And The Role Of Gln157
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-06-20 Classification: HYDROLASE Ligands: ZN, CA, MG, ACT |

