Search Count: 19
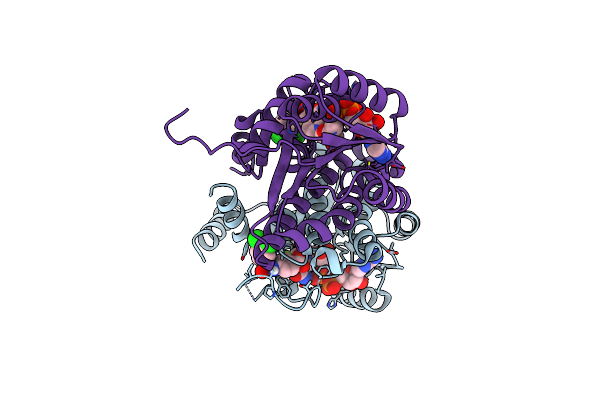 |
Design And Application Of Synthetic 17B-Hsd13 Substrates To Drug Discovery, And To Reveal Preserved Catalytic Activity Of Protective Human Variants
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-11-27 Classification: HYDROLASE Ligands: NAD, A1AG4 |
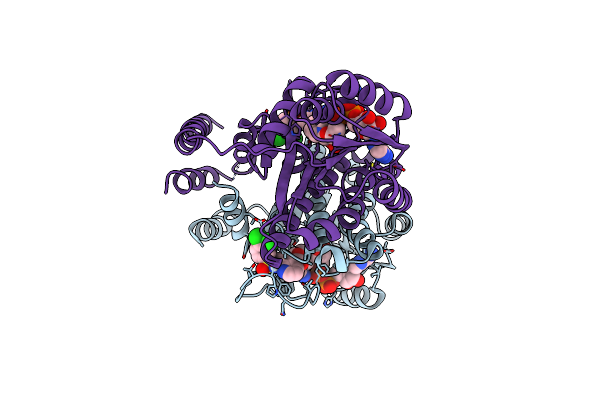 |
Design And Application Of Synthetic 17B-Hsd13 Substrates To Drug Discovery, And To Reveal Preserved Catalytic Activity Of Protective Human Variants
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2024-11-27 Classification: HYDROLASE Ligands: NAD, A1AG5 |
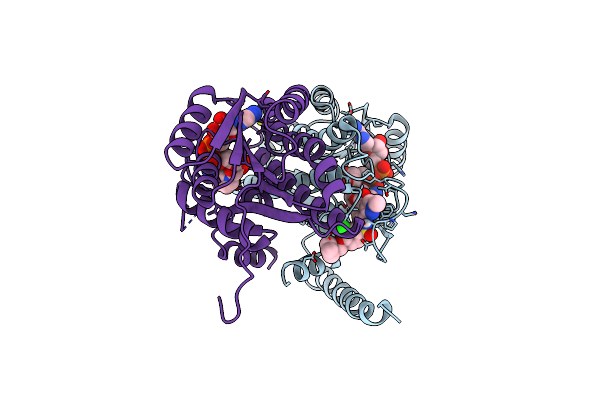 |
Design And Application Of Synthetic 17B-Hsd13 Substrates To Drug Discovery, And To Reveal Preserved Catalytic Activity Of Protective Human Variants
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-11-27 Classification: HYDROLASE Ligands: NAD, A1AG6 |
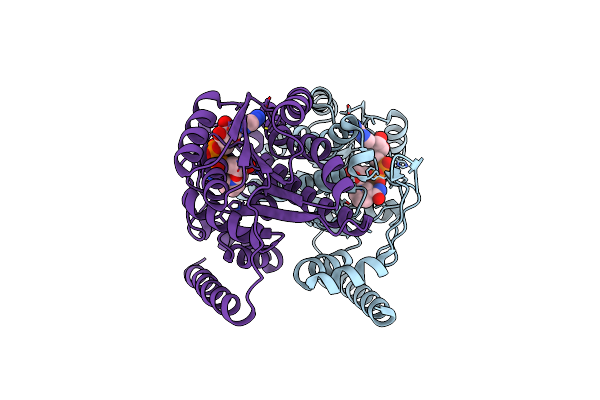 |
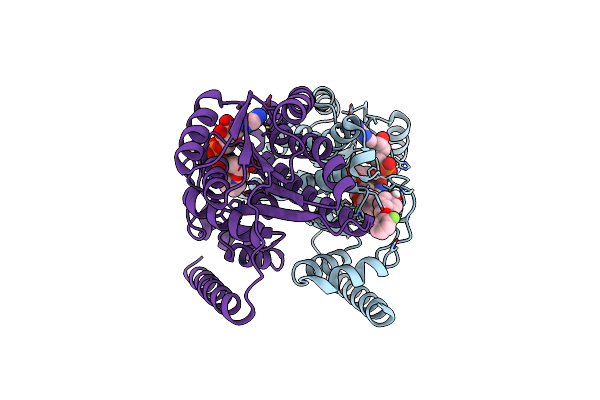
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2023-08-09 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
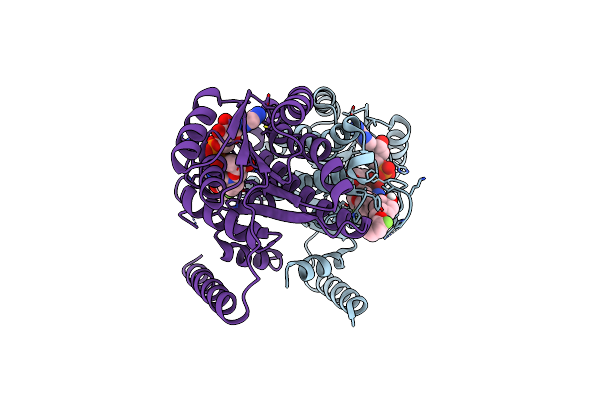
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2023-08-09 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAD, YXW |
 |
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-08-09 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAD, YXW |
 |
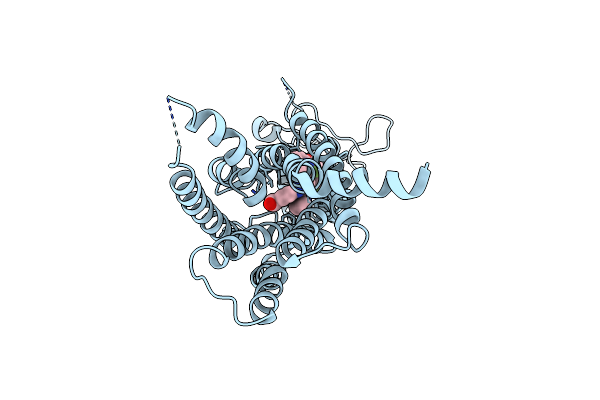
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-08-09 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAD, YYC, CE1, SO4 |
 |
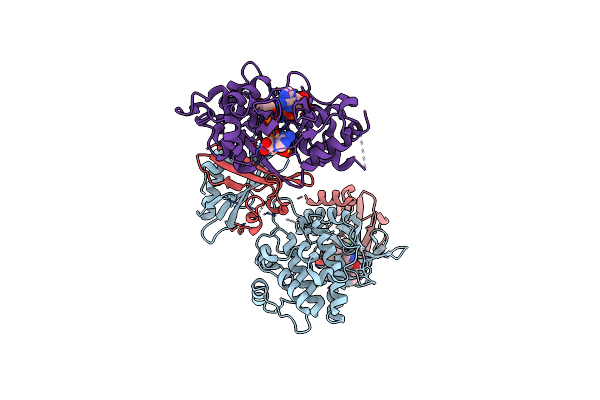
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: MEMBRANE PROTEIN Ligands: 82L |
 |
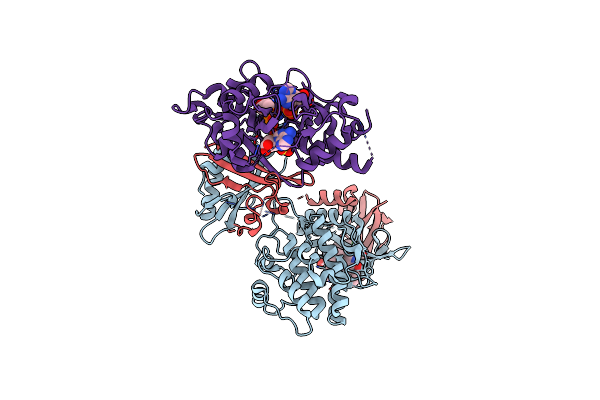
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-08-08 Classification: TRANSFERASE Ligands: HTV, STU, CL, AMP, ADP, SO4 |
 |
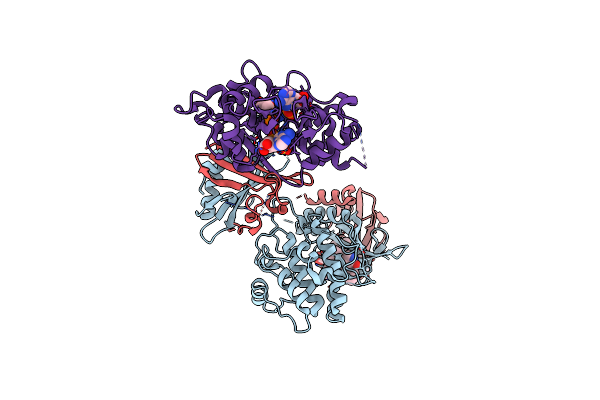
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.27 Å Release Date: 2018-08-08 Classification: TRANSFERASE Ligands: STU, HU7, CL, AMP, ADP, SO4 |
 |
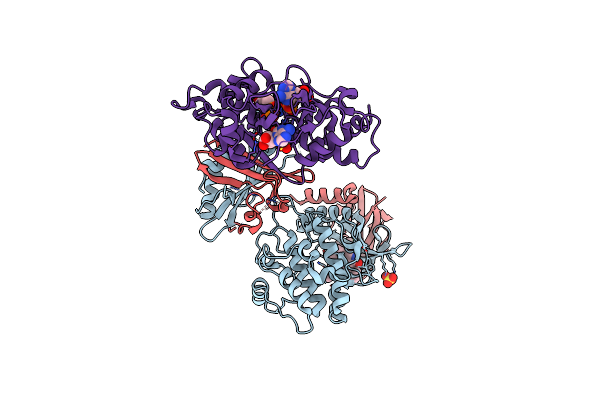
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2018-08-08 Classification: TRANSFERASE Ligands: STU, HUG, CL, AMP, ADP, SO4 |
 |
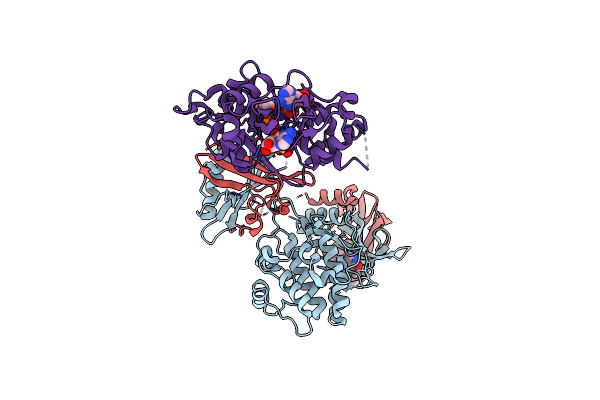
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.46 Å Release Date: 2017-03-29 Classification: TRANSFERASE Ligands: STU, CL, SO4, 75O, AMP, ADP |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.41 Å Release Date: 2016-08-17 Classification: TRANSFERASE Ligands: STU, 6VT, CL, AMP, ADP, SO4 |
 |
 |
Organism: Heloderma suspectum, conus planorbis
Method: SOLUTION NMR Release Date: 2016-05-04 Classification: TOXIN |
 |
 |
Nmr Structure Of A Short Hydrophobic 11Mer Peptide In Dmso-D6/H2O (1:3) Solution
|
 |
 |

