Search Count: 13
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-16 Classification: ANTIVIRAL PROTEIN |
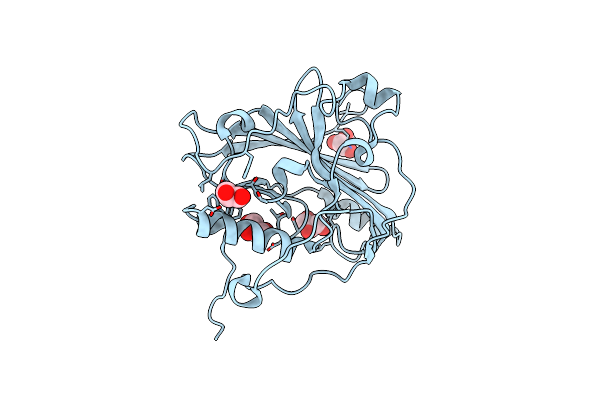 |
Organism: Equid alphaherpesvirus 4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-11-11 Classification: VIRAL PROTEIN Ligands: GOL |
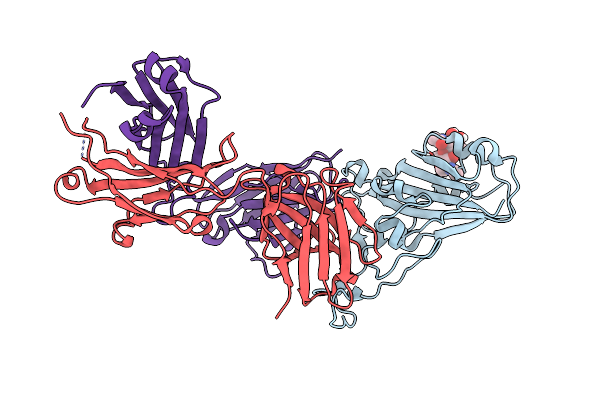
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Antibody Cv07-270
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2020-10-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, SO4 |
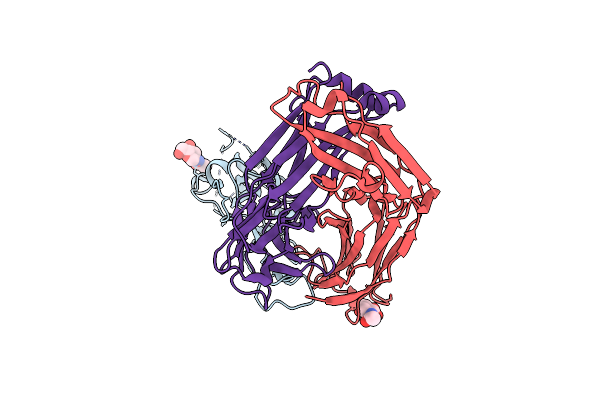 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Antibody Cv07-250
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2020-10-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Equid alphaherpesvirus 1
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-09-30 Classification: VIRAL PROTEIN Ligands: MG, NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: MN, EDO, GOL, SO4, 16P |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: EDO, CL, MN |
 |
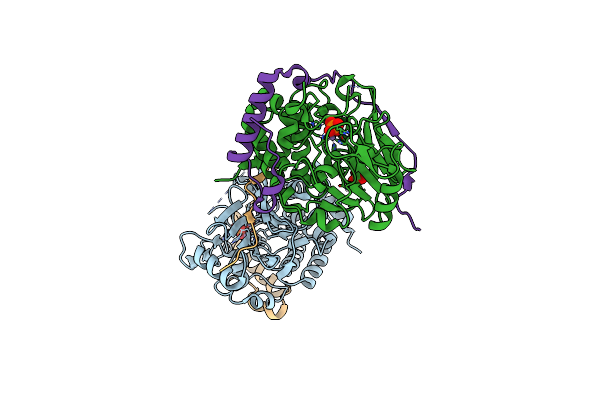
Structure Of Pp1-Irsp53 Chimera [Pp1(7-304) + Linker (G/S)X9 + Irsp53(449-465)] Bound To Phactr1 (516-580)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: MN, PO4, EDO, 16P |
 |
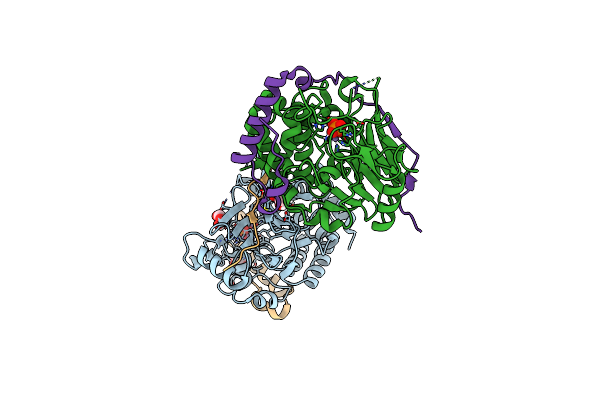
Structure Of Pp1-Spectrin Alpha Ii Chimera [Pp1(7-304) + Linker (G/S)X9 + Spectrin Alpha Ii (1025-1039)] Bound To Phactr1 (516-580)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: MN, PO4, SO4 |
 |
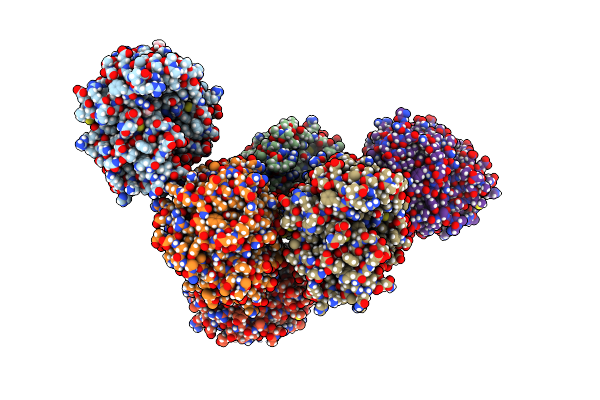
Structure Of Pp1-Irsp53 S455E Chimera [Pp1(7-304) + Linker (G/S)X9 + Irsp53(449-465)] Bound To Phactr1 (516-580)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: MN, GOL, PO4 |
 |
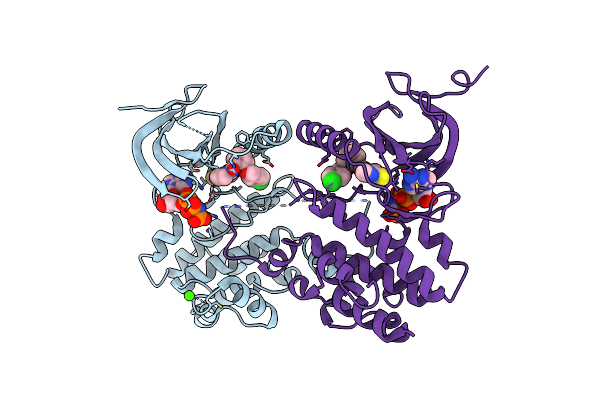
Structure Of Pp1-Phactr1 Chimera [Pp1(7-304) + Linker (Sgsgs) + Phactr1(526-580)]
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: MN, GOL, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-10-19 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 7AV, ANP, MN, CA |
 |
Crystal Structure Of ((2S)-5-Amino-2-((1-N-Propyl-1H-Imidazol-4-Yl) Methyl)Pentanoic Acid) Uk396,082 A Tafia Inhibitor, Bound To Activated Porcine Pancreatic Carboxypeptidaseb
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2007-11-20 Classification: HYDROLASE Ligands: ZN, 720 |

