Search Count: 61
 |
Organism: Nepenthes alata x nepenthes ventricosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-10-22 Classification: HYDROLASE Ligands: SO4, CL, TRS, PEG, GOL, GLY |
 |
Crystal Structure Of Thomasclavelia Ramosa Iga Peptidase (Igase) Active Site Mutant (E330-N876)
Organism: Thomasclavelia ramosa
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-06-25 Classification: HYDROLASE Ligands: ZN, PG4, EDO, FMT |
 |
Cryo-Em Structure Of Thomasclavelia Ramosa Iga Peptidase (Igase) Active Site Mutant (S32-N876)
Organism: Thomasclavelia ramosa
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: HYDROLASE Ligands: ZN, GOL, AZI |
 |
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-02-28 Classification: PROTEIN TRANSPORT Ligands: CL, PEG, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2023-03-22 Classification: DNA BINDING PROTEIN Ligands: DHT, EDO |
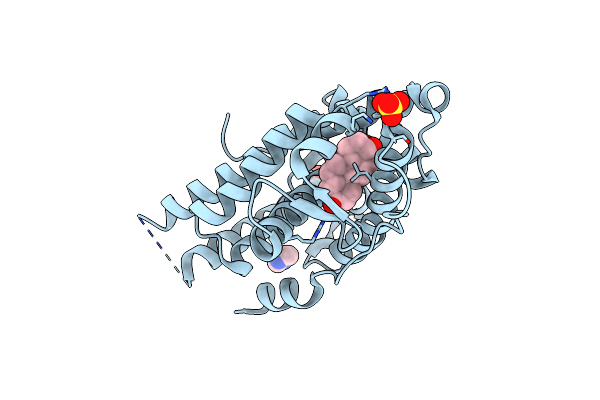 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2023-03-22 Classification: DNA BINDING PROTEIN Ligands: DHT, SO4, IMD |
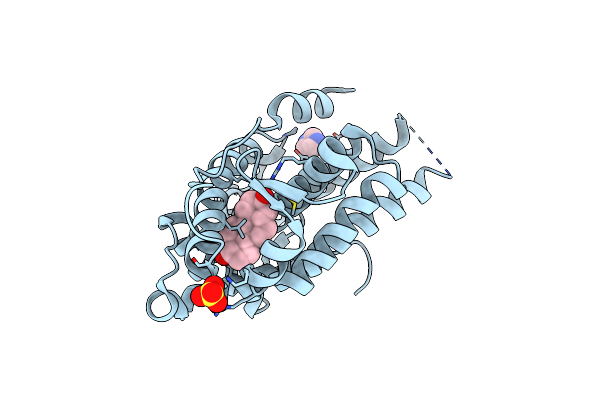 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-03-22 Classification: DNA BINDING PROTEIN Ligands: DHT, IMD, SO4 |
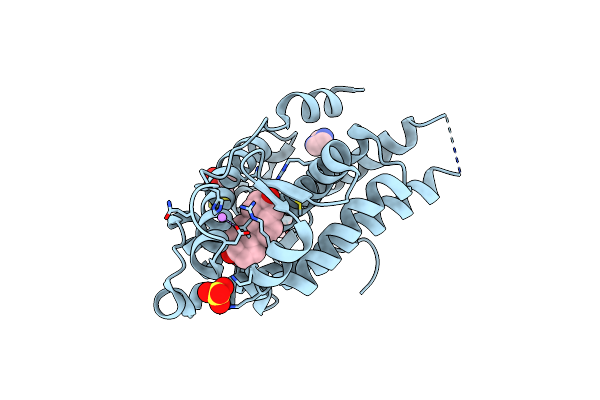 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2023-03-22 Classification: DNA BINDING PROTEIN Ligands: DHT, GOL, SO4, DTT, IMD, LI |
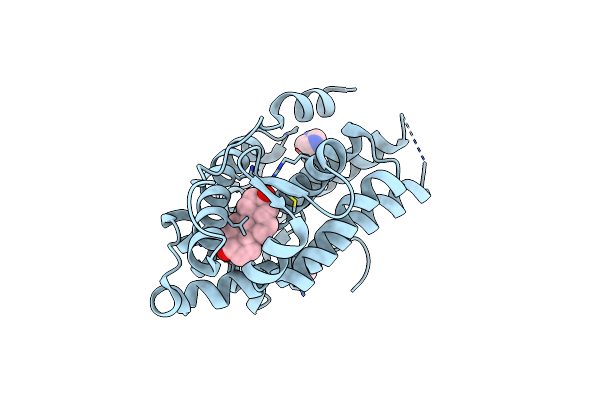 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-03-22 Classification: DNA BINDING PROTEIN Ligands: DHT, IMD |
 |
Organism: Methanosarcina acetivorans c2a, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-03-22 Classification: HYDROLASE Ligands: CA, ZN, AES, GOL |
 |
Organism: Methanosarcina acetivorans c2a
Method: X-RAY DIFFRACTION Resolution:4.50 Å Release Date: 2023-03-22 Classification: HYDROLASE Ligands: ZN, CA |
 |
Matrix-Metallopeptidase Inhibitor Potempin A (Pota) From Tannerella Forsythia
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR Ligands: NI |
 |
Potempin A (Pota) From Tannerella Forsythia In Complex With The Catalytic Domain Of Human Mmp-12
Organism: Homo sapiens, Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR Ligands: CA, ZN |
 |
Matrix-Metallopeptidase Inhibitor Potempin A (Pota) From Tannerella Forsythia In Complex With T. Forsythia Karilysin.
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR Ligands: ZN, CA, MES, GOL |
 |
Structure Of Tannerella Forsythia Selenomethionine-Derivatized Potempin D Mutant I53M
Organism: Tannerella forsythia (strain atcc 43037 / jcm 10827 / ccug 21028 a / kctc 5666 / fdc 338)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR Ligands: EDO |
 |
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR Ligands: GOL |
 |
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR Ligands: EPE |
 |
Organism: Tannerella forsythia
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2022-12-21 Classification: HYDROLASE INHIBITOR |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-10-12 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-10-12 Classification: DE NOVO PROTEIN Ligands: MG |

