Search Count: 28
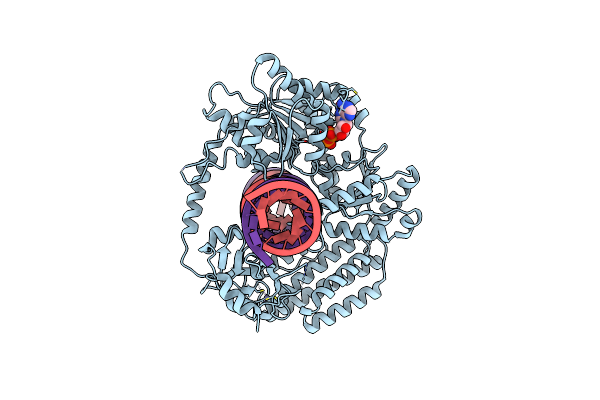 |
Organism: Caenorhabditis elegans, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: ANTIVIRAL PROTEIN/RNA Ligands: ZN, MG, ADP |
 |
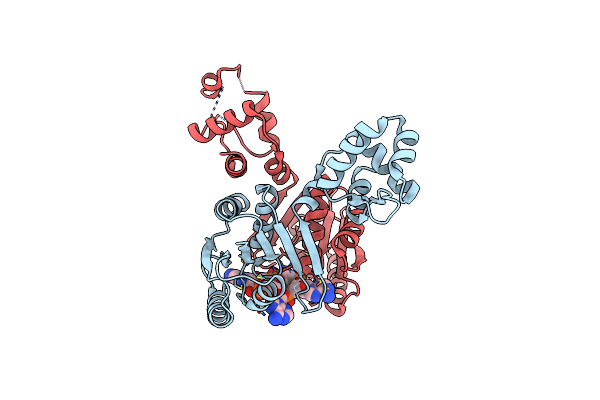
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-31 Classification: SIGNALING PROTEIN Ligands: CA |
 |
The Structure Of Sulfolobus Solfataricus Csa3 In Complex With Cyclic Tetraadenylate (Ca4)
Organism: Saccharolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2), Saccharolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2021-03-10 Classification: TRANSCRIPTION |
 |
Organism: Saccharomyces cerevisiae (strain yjm789)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-11-25 Classification: SIGNALING PROTEIN Ligands: HEM |
 |
Pie12 D-Peptide Against Hiv Entry (In Complex With Iqn17 Q577R Resistance Mutant)
Organism: Synthetic construct, Saccharomyces cerevisiae, Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-02-05 Classification: VIRAL PROTEIN/INHIBITOR Ligands: CL |
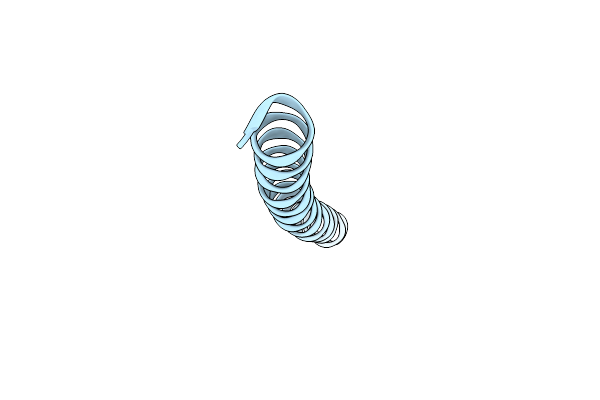 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-10-22 Classification: BIOSYNTHETIC PROTEIN |
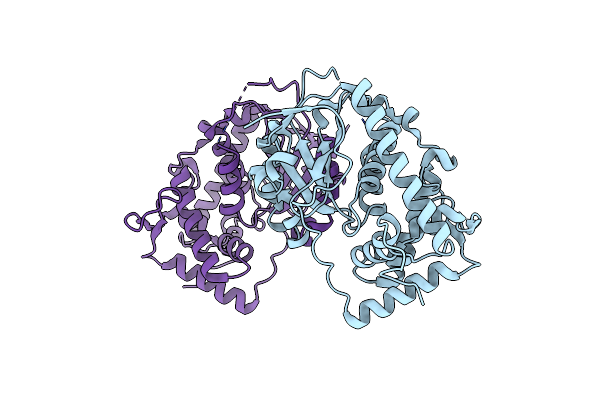 |
Crystal Structure Of Sfh3, A Phosphatidylinositol Transfer Protein That Integrates Phosphoinositide Signaling With Lipid Droplet Metabolism
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2014-09-10 Classification: SIGNALING PROTEIN |
 |
Organism: Acidianus hospitalis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2013-11-06 Classification: PROTEIN TRANSPORT Ligands: CL |
 |
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2013-11-06 Classification: HYDROLASE Ligands: CL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2012-09-05 Classification: HYDROLASE/CHAPERONE Ligands: MG, LDZ |
 |
Method: X-RAY DIFFRACTION
Resolution:1.40 Å Release Date: 2011-03-02 Classification: VIRAL PROTEIN/VIRAL PROTEIN inhibitor |
 |
Method: X-RAY DIFFRACTION
Resolution:1.55 Å Release Date: 2010-11-03 Classification: DE NOVO PROTEIN |
 |
Method: X-RAY DIFFRACTION
Resolution:1.45 Å Release Date: 2010-11-03 Classification: DE NOVO PROTEIN Ligands: CXS |
 |
Method: X-RAY DIFFRACTION
Resolution:1.45 Å Release Date: 2010-11-03 Classification: DE NOVO PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-10-27 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2010-10-13 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-07-21 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-06-30 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2009-06-30 Classification: PROTEIN BINDING Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2009-06-30 Classification: PROTEIN TRANSPORT |

