Search Count: 148
 |
Crystal Structures Of 5-Aminoimidazole Ribonucleotide (Air) Synthetase, Purm, From Thermus Thermophilus
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-11-25 Classification: LIGASE Ligands: SO4 |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-01-14 Classification: TRANSFERASE/DNA Ligands: MG, 1FZ |
 |
Organism: Japanese encephalitis virus
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2014-12-24 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of Hypothetical Protein Pf0907 From Pyrococcus Furiosus Solved By Sulfur Sad Using Swiss Light Source Data
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-10 Classification: UNKNOWN FUNCTION Ligands: CL |
 |
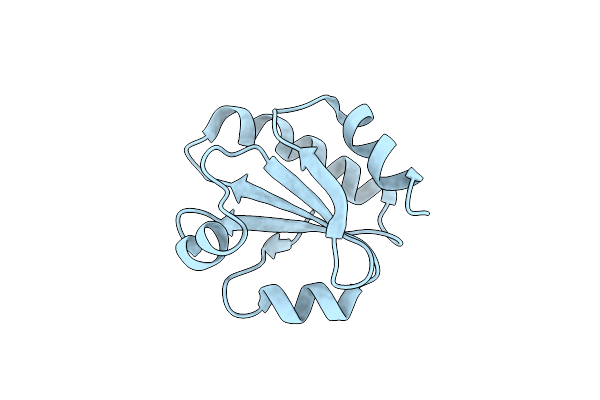
Crystal Structure Of Hypothetical Protein Pf0907 From Pyrococcus Furiosus Solved By Sulfur Sad Using Swiss Light Source Data
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2014-12-10 Classification: UNKNOWN FUNCTION Ligands: CL, IMD |
 |
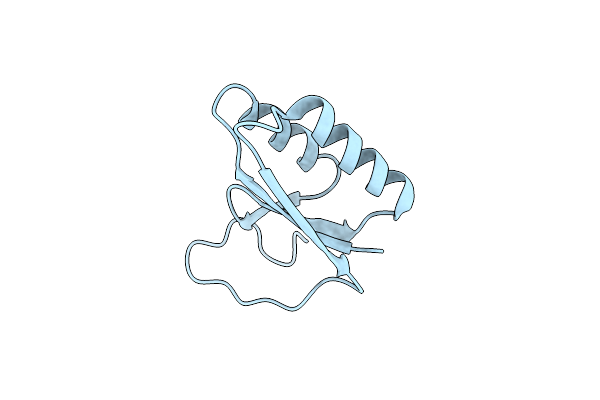
Crystal Structure Of Thermus Thermophilus Thioredoxin Solved By Sulfur Sad Using Swiss Light Source Data
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-12-10 Classification: ELECTRON TRANSPORT Ligands: CL |
 |
Hypothetical Protein Pf1117 From Pyrococcus Furiosus: Structure Solved By Sulfur-Sad Using Swiss Light Source Data
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2014-12-10 Classification: HYDROLASE Ligands: CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-12-10 Classification: ISOMERASE Ligands: GSH, LVJ |
 |
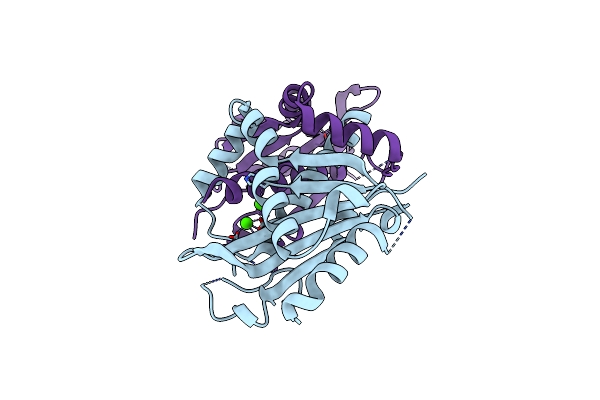
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-12-10 Classification: CELL CYCLE |
 |
Crystal Structure Of The Exonuclease Domain Of Qip (Qde-2 Interacting Protein) Solved By Native-Sad Phasing.
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2014-12-10 Classification: RNA BINDING PROTEIN Ligands: CA |
 |
Conserved Hypothetical Protein Pf1771 From Pyrococcus Furiosus Solved By Sulfur Sad Using Swiss Light Source Data
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-10 Classification: OXIDOREDUCTASE |
 |
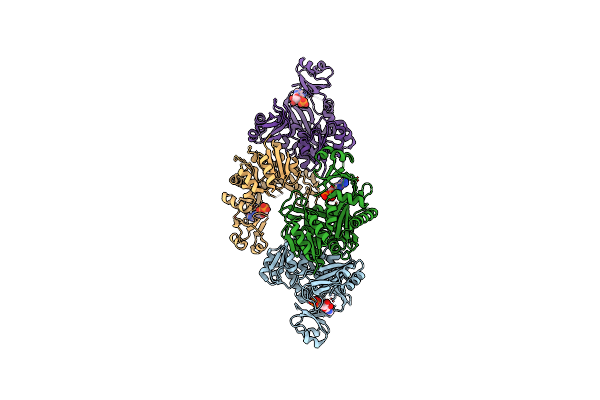
Crystal Structure Of Formyltetrahydrofolate Deformylase From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2014-01-08 Classification: HYDROLASE |
 |
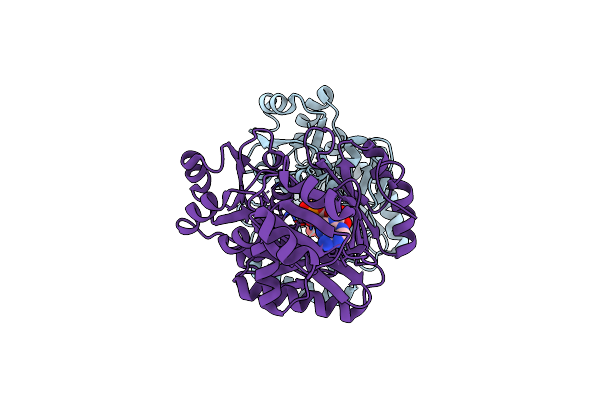
Crystal Structure Of N5-Carboxyaminoimidazole Ribonucleotide Synthetase From Thermotoga Maritima
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-04-25 Classification: LIGASE Ligands: ADP |
 |
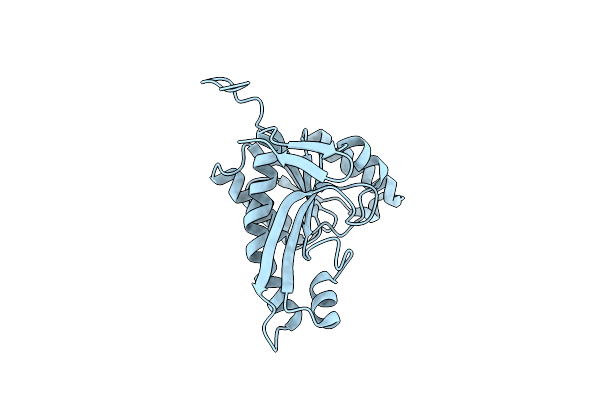
Crystal Structure Of N5-Carboxyaminoimidazole Ribonucleotide Synthetase From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-04-11 Classification: LIGASE Ligands: AMP, CL |
 |
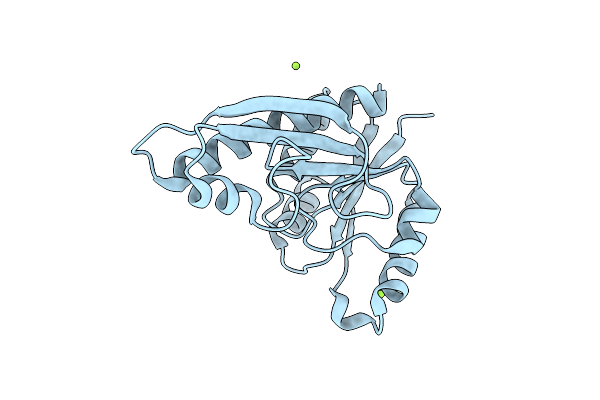
Crystal Structure Of Glycinamide Ribonucleotide Transformylase 1 From Symbiobacterium Toebii
Organism: Symbiobacterium toebii
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2012-03-07 Classification: TRANSFERASE |
 |
Crystal Structure Of Glycinamide Ribonucleotide Transformylase 1 From Geobacillus Kaustophilus
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-03-07 Classification: TRANSFERASE Ligands: MG |
 |
Crystal Structure Of The Hypothetical Tandem-Type Universal Stress Protein Ttha0350 From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2010-12-15 Classification: UNKNOWN FUNCTION |
 |
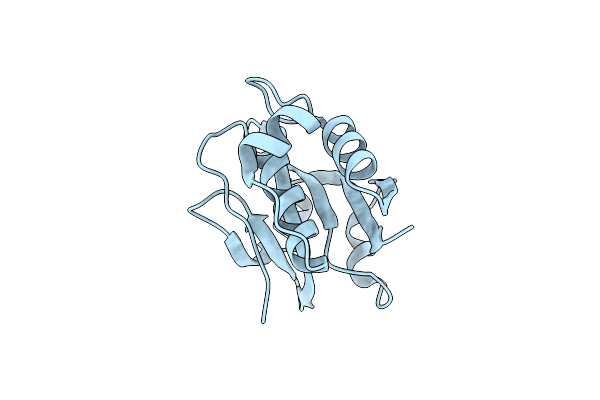
Crystal Structure Of The Hypothetical Tandem-Type Universal Stress Protein Ttha0350 Complexed With Atps.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-12-15 Classification: UNKNOWN FUNCTION Ligands: ATP, MG |
 |
Crystal Structure Of The Peptidase Domain Of Streptococcus Coma, A Bi-Functional Abc Transporter Involved In Quorum Sensing Pathway
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-02-23 Classification: HYDROLASE |
 |
Crystal Structure Of Hypoxanthine-Guanine Phosphoribosyltransferase From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2010-02-09 Classification: TRANSFERASE Ligands: DIO |

