Search Count: 38,702
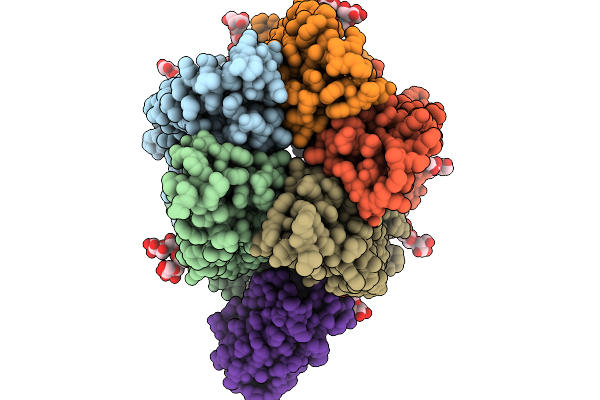 |
A5B3 Gabaar Bound To Gaba And Mb25 In A Desensitized State In Saposin Nanodiscs After Short Gaba Treatment
Organism: Aequorea victoria, Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: ABU, EPE, NAG |
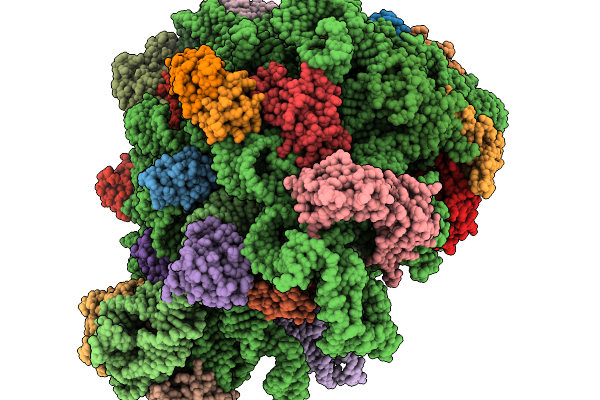 |
Structure Of Wt E.Coli Ribosome With Complexed Filament Nascent Chain At Length 47, With P-Site Trna
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME |
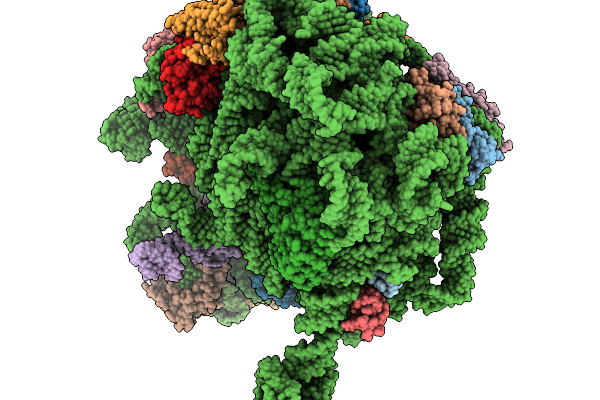 |
Structure Of Wt E.Coli Ribosome With Complexed Filament Nascent Chain At Length 31, With P-Site Trnas
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME |
 |
Structure Of Undecaprenyl-Phosphate 4-Deoxy-4-Formamido-L-Arabinose Transferase Embedded In Nanodisc
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: ELECTRON TRANSPORT Ligands: HEO, HEM, CU, ZN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2026-01-14 Classification: TRANSFERASE Ligands: GOL, MN, AMP, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-01-14 Classification: TRANSFERASE Ligands: AMP, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2026-01-14 Classification: LYASE Ligands: NAD, SO4 |
 |
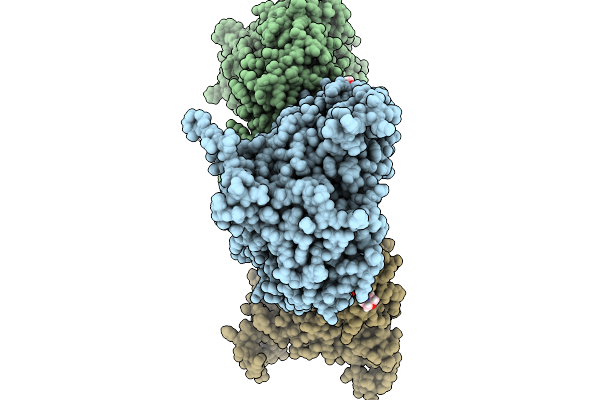
Hexamer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Resolution:2.87 Å Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.31 Å Release Date: 2026-01-14 Classification: IMMUNE SYSTEM Ligands: MES |
 |
Crystal Structure Of Engineered Glutamine Binding Protein And A Gd-Dota Ligand - Gln Bound
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2026-01-14 Classification: METAL BINDING PROTEIN Ligands: GLN, ACT, GOL, A1C21, GD3, SO4 |
 |
Crystal Structure Of Engineered Glutamine Binding Protein And A Gd-Dota Ligand - No Gln Bound
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2026-01-14 Classification: METAL BINDING PROTEIN Ligands: SO4, ACT, GOL, A1C21, EDO, GD3 |
 |
Structures And Mechanism Of The Nicotinamide Nucleotide Transhydrogenase From E. Coli
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: PC1, PKZ |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: CL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: CL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: CL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: CL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:2.60 Å Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN/RNA/DNA Ligands: ATP, MG |
 |
Hexamer Msp1 From S.Cerevisiae(With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |

