Search Count: 1,70,810
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: PROTEIN BINDING Ligands: ACT, EDO |
 |
Nadph And 1-Benzyl-4-Methylpiperidin-3-One Complex Structure Of Imine Reductase Wild Type From Pochonia Chlamydosporia 170
Organism: Pochonia chlamydosporia 170
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: OXIDOREDUCTASE Ligands: NAP, TRS, PEG, A1D7O |
 |
Nadph And 1-Benzyl-4-Methylpiperidin-3-One Complex Structure Of Imine Reductase Mutant(M6) From Pochonia Chlamydosporia 170
Organism: Pochonia chlamydosporia 170
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: OXIDOREDUCTASE Ligands: NAP, PEG, A1D7O |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, F3S, SAM, YVI, PO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: FLUORESCENT PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: FLUORESCENT PROTEIN Ligands: GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: FLUORESCENT PROTEIN |
 |
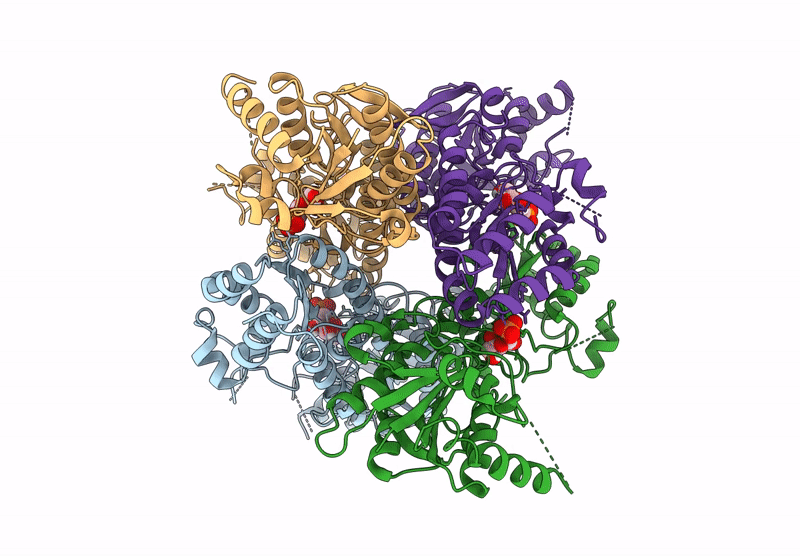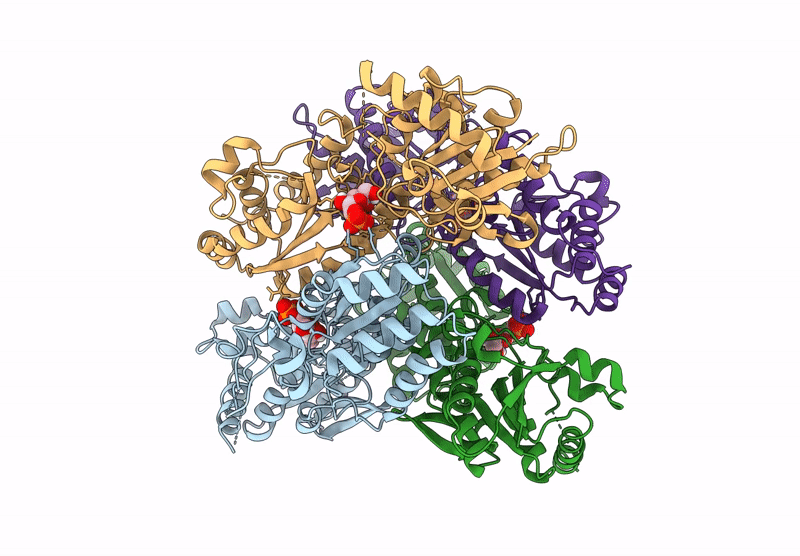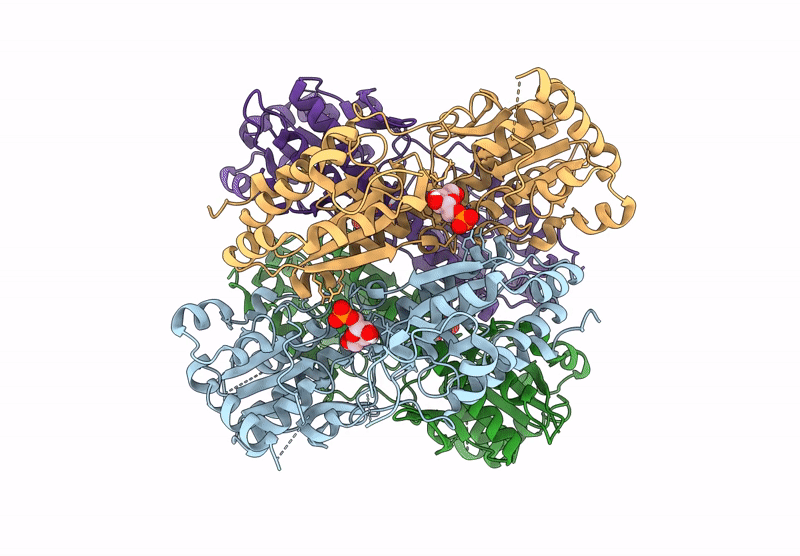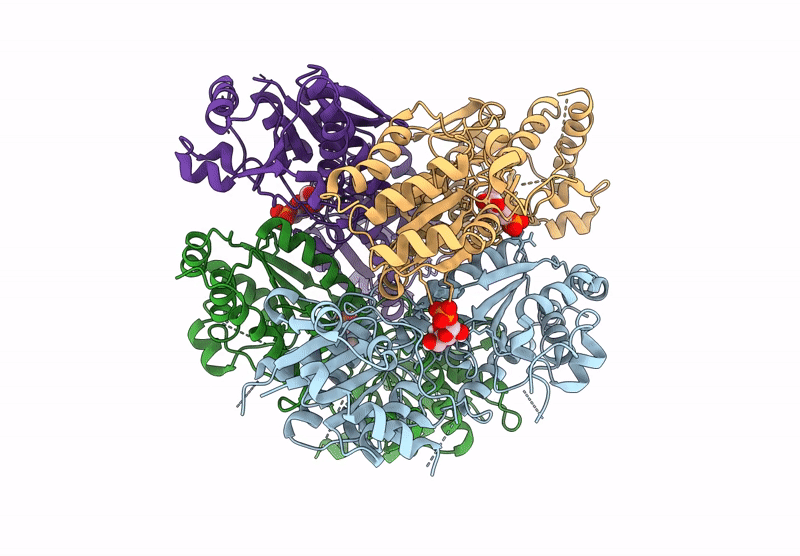
Crystal Structure Of Pyrophosphate-Dependent Phosphofructokinase From Candidatus Prometheoarchaeum Syntrophicum
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE |
 |
Crystal Structure Of The Pyrophosphate-Dependent Phosphofructokinase From Candidatus Prometheoarchaeum Syntrophicum With Fructose 6-Phosphate
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: F6P |
 |
Crystal Structure Of Pyrophosphate-Dependent Phosphofructokinase From Candidatus Prometheoarchaeum Syntrophicum With Phosphoenolpyruvate, Phosphate And Mg2+
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: PEP, PO4, MG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE |
 |
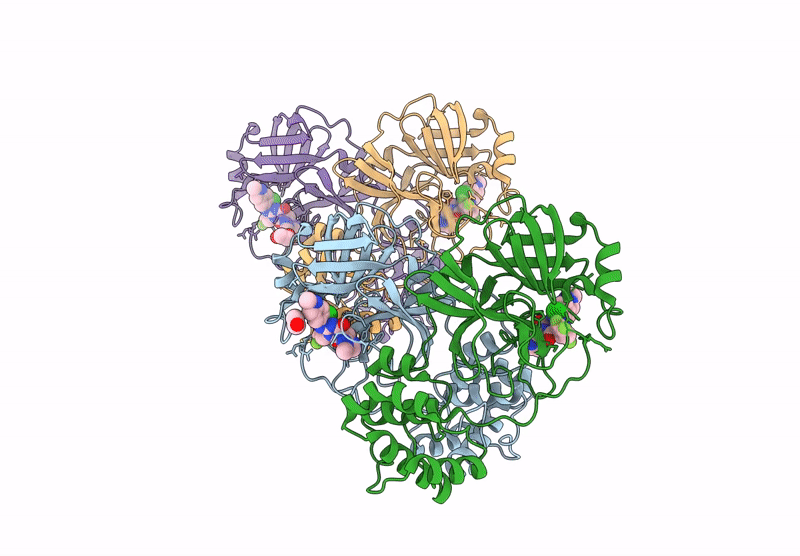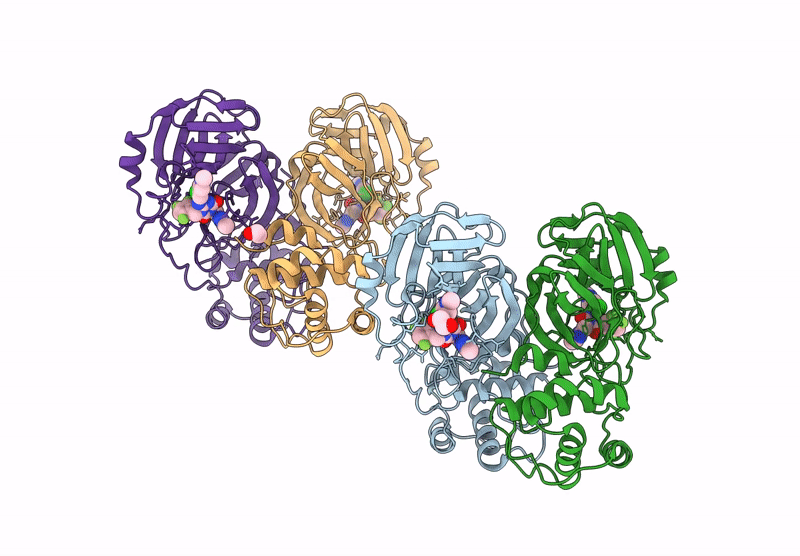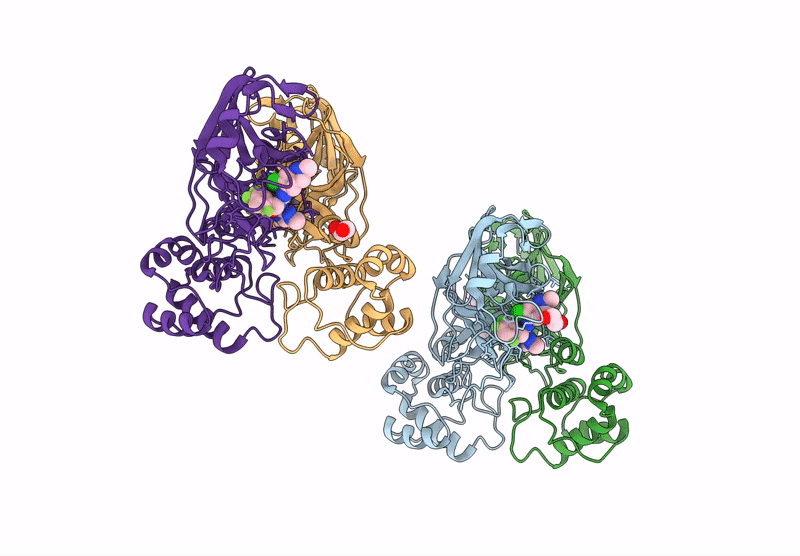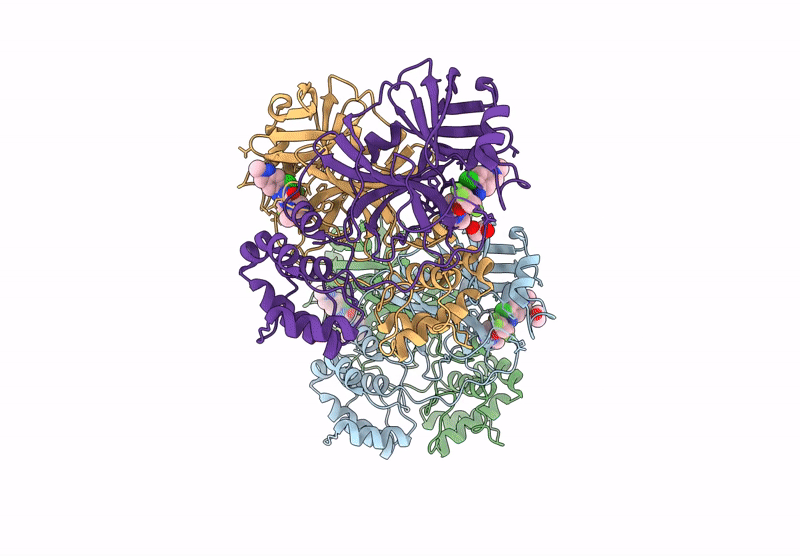
X-Ray Crystal Structure Of Sars-Cov-2 Main Protease Quadruple Mutants In Complex With Nirmatrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: 4WI, EDO, BR, SO4 |
 |
X-Ray Crystal Structure Of Sars-Cov-2 Main Protease Quadruple Mutants In Complex With Ensitrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: 7YY, EDO |
 |
X-Ray Crystal Structure Of Sars-Cov-2 Main Protease Double Mutants In Complex With Ensitrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: 7YY, MG, CL |
 |
X-Ray Crystal Structure Of Sars-Cov-2 Main Protease Triple Mutants In Complex With Bofutrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: FHR |
 |
X-Ray Crystal Structure Of Sars-Cov-2 Main Protease Quadruple Mutants In Complex With Bofutrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: FHR |
 |
Structure Of Sars-Cov-2 Main Protease In Complex With Bofutrelvir In Orthorhombic Form
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: FHR, EDO, PEG |
 |
X-Ray Crystal Structure Of Sars-Cov-2 Main Protease Complex With Bofutrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: FHR, DMS |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE Ligands: ZN, MCO |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE Ligands: ZN, X8Z, ACT |

