Search Count: 45
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Macrolide Erythromycin, Mrna, Aminoacylated A-Site Lys-Trnalys, P-Site Fmrc-Peptidyl-Trnamet, And Deacylated E-Site Trnalys At 2.65A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: RIBOSOME Ligands: MG, K, ERY, ZN, SF4 |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Macrolide Erythromycin, Mrna, Aminoacylated A-Site Lys-Trnalys, P-Site Fmac-Peptidyl-Trnamet, And Deacylated E-Site Trnalys At 2.70A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: RIBOSOME Ligands: MG, K, ERY, ZN, SF4 |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Macrolide Erythromycin, Mrna, Deacylated A-Site Trnaphe, P-Site Fmrc-Peptidyl-Trnamet, And Deacylated E-Site Trnaphe At 2.75A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: RIBOSOME Ligands: MG, K, ERY, ZN, SF4 |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx And Erythromycin:50S-Hflx-B-Ery
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: RIBOSOME Ligands: GCP, ERY |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx And Erythromycin:50S-Hflx-A-Ery
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP, ERY |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: RIBOSOME Ligands: ERY |
 |
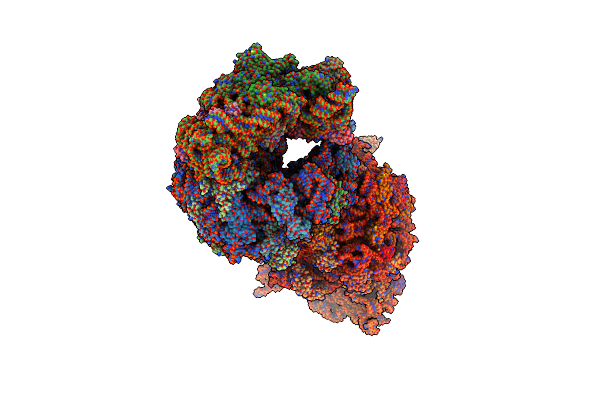
Crystal Structure Of The A2058-N6-Dimethylated Thermus Thermophilus 70S Ribosome In Complex With Protein Y, Hygromycin A, And Erythromycin At 2.45A Resolution
Organism: Escherichia coli k-12, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-07-26 Classification: RIBOSOME Ligands: MG, HGR, ERY, MPD, ARG, ZN, SF4 |
 |
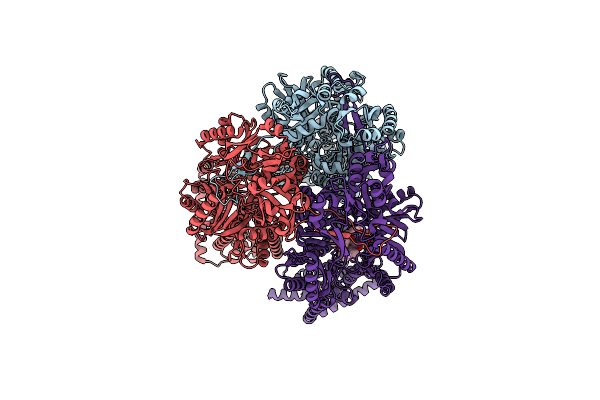
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, Hygromycin A, And Erythromycin At 2.50A Resolution
Organism: Escherichia coli k-12, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-12 Classification: RIBOSOME Ligands: MG, HGR, ERY, MPD, ARG, ZN, SF4 |
 |
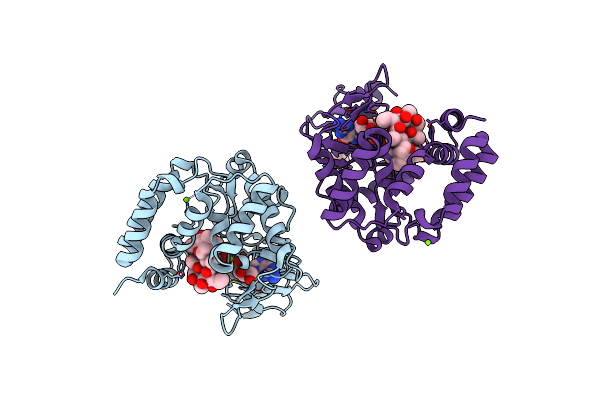
Organism: Campylobacter jejuni
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: MEMBRANE PROTEIN Ligands: ERY |
 |
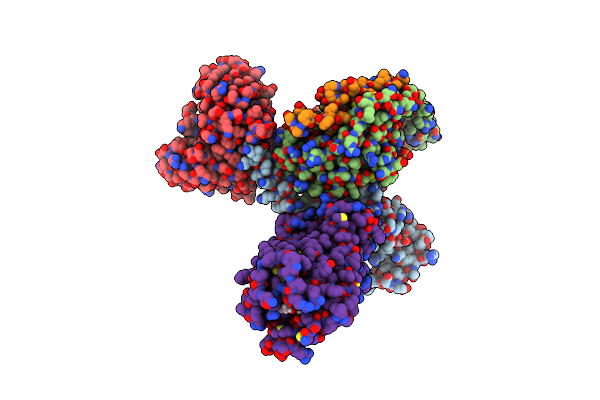
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2023-04-26 Classification: TRANSFERASE Ligands: ERY, GTP, MG, GOL |
 |
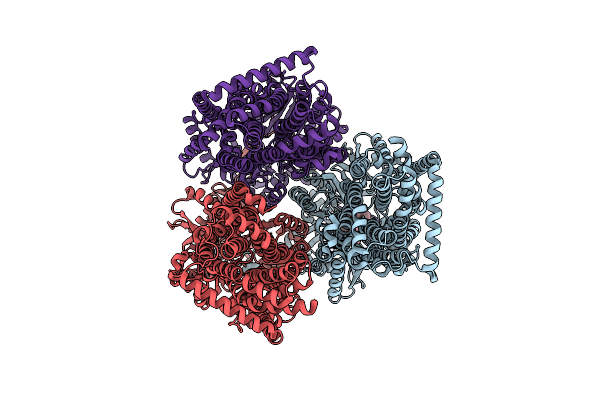
Cryo-Em Structure Of The Erythromycin-Bound Motilin Receptor-Gq Protein Complex
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: SIGNALING PROTEIN Ligands: ERY |
 |
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: MEMBRANE PROTEIN Ligands: ERY |
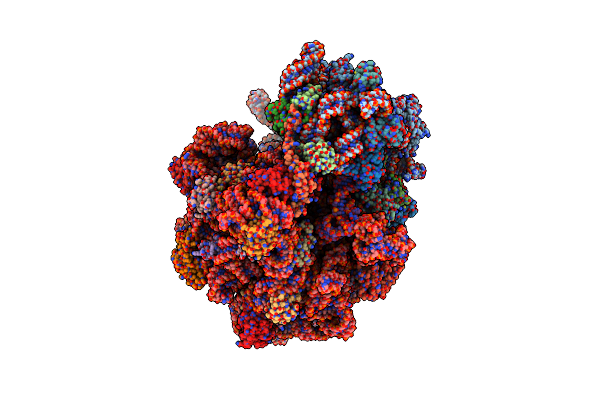 |
Erythromycin-Stalled Escherichia Coli 70S Ribosome With Streptococcal Msrdl Nascent Chain
Organism: Escherichia coli, Escherichia coli k-12, Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: RIBOSOME Ligands: MG, ERY |
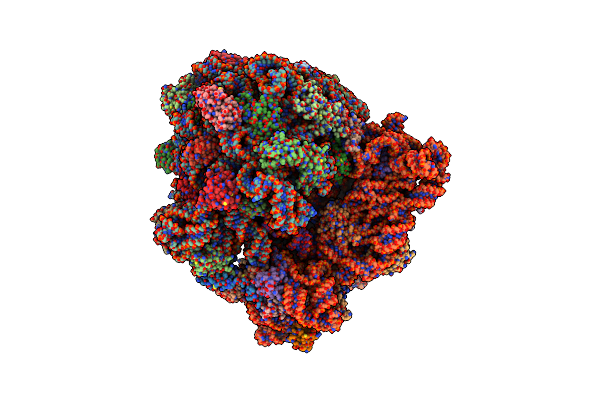 |
Structure Of Ermdl-Erythromycin-Stalled 70S E. Coli Ribosomal Complex With A And P-Trna
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-08-11 Classification: RIBOSOME Ligands: ERY |
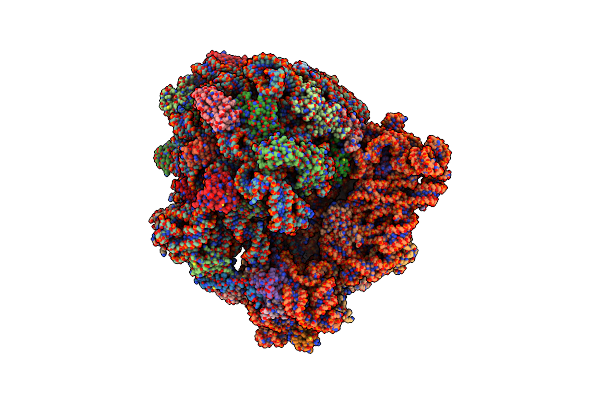 |
Structure Of Ermdl-Erythromycin-Stalled 70S E. Coli Ribosomal Complex With P-Trna
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-07-14 Classification: RIBOSOME Ligands: ERY |
 |
E. Coli 70S Containing Suppressor Trna In The A-Site Stabilized By A Negamycin Analogue And P-Site Trna-Nascent Chain.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: RIBOSOME Ligands: MG, ERY, SY5 |
 |
Erythromycin Binding To The Access Pocket Of Acrb-G616P L Protomer And 3-Formylrifamycin Sv Binding To The Access Pocket Of Acrb-G616P T Protomer
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2021-05-19 Classification: TRANSPORT PROTEIN Ligands: C14, LMT, ERY, EDO, GOL, D12, OCT, HEX, 3YI, DDQ, SO4, PG4, PTY |
 |
Crystal Structure Of The A2058-Unmethylated Thermus Thermophilus 70S Ribosome In Complex With Erythromycin And Protein Y (Yfia) At 2.55A Resolution
Organism: Escherichia coli (strain k12), Thermus thermophilus hb8, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2020-12-23 Classification: RIBOSOME Ligands: MG, ERY, MPD, ARG, ZN, SF4 |
 |
Cryo-Electron Microscopy Structures Of A Gonococcal Multidrug Efflux Pump Illuminate A Mechanism Of Erythromycin Drug Recognition
Organism: Neisseria gonorrhoeae
Method: ELECTRON MICROSCOPY Release Date: 2020-07-01 Classification: MEMBRANE PROTEIN Ligands: PTY, ERY |
 |
Erythromycin Resistant Staphylococcus Aureus 70S Ribosome (Delta R88 A89 Ul22) In Complex With Erythromycin.
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2019-08-21 Classification: RIBOSOME Ligands: ERY |

