Search Count: 13
 |
Crystal Structure Of Udp-N-Acetylenolpyruvoylglucosamine Reductase (Murb) From Brucella Ovis
Organism: Brucella ovis atcc 25840
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2025-01-22 Classification: OXIDOREDUCTASE Ligands: SO4, FAD, EPU |
 |
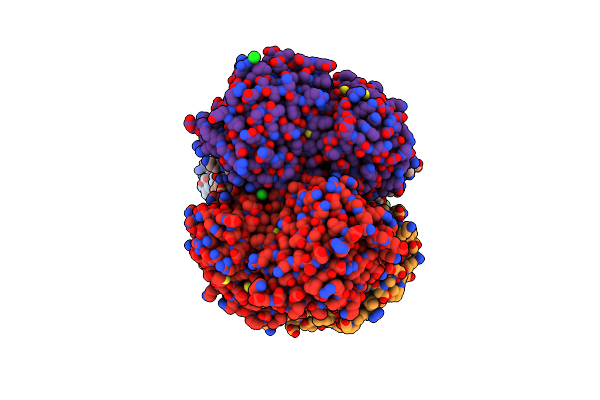
Crystal Structure Of Mura From Clostridium Difficile, Mutation C116S, In The Presence Of Uridine-Diphosphate-2(N-Acetylglucosaminyl) Butyric Acid
Organism: Peptoclostridium difficile (strain 630)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-11-27 Classification: TRANSFERASE Ligands: EPU, EDO, NA |
 |
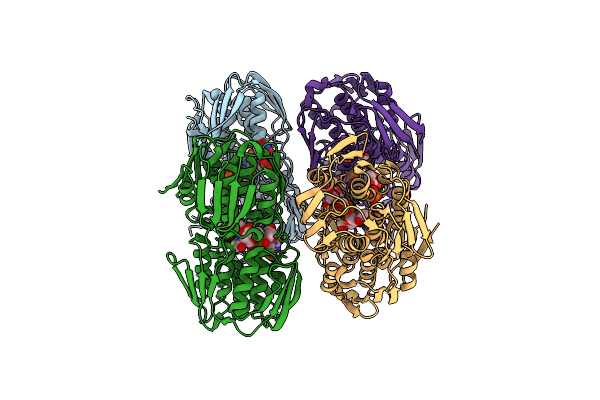
2.75 Angstrom Resolution Crystal Structure Of Udp-N-Acetylglucosamine 1-Carboxyvinyltransferase From Pseudomonas Putida In Complex With Uridine-Diphosphate-2(N-Acetylglucosaminyl) Butyric Acid, (2R)-2-(Phosphonooxy)Propanoic Acid And Magnesium
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2018-03-21 Classification: TRANSFERASE Ligands: EPU, 0V5, MG, CL |
 |
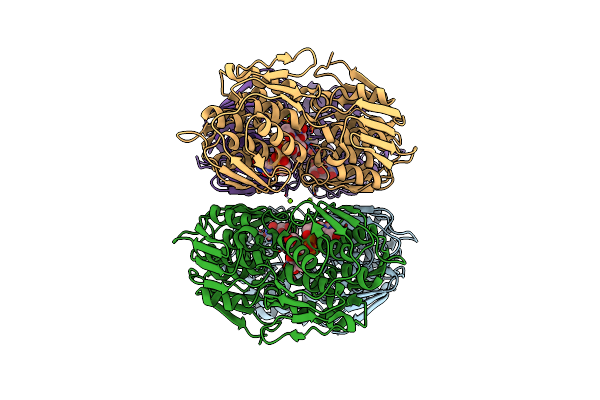
2.0 Angstrom Resolution Crystal Structure Of Udp-N-Acetylglucosamine 1-Carboxyvinyltransferase From Streptococcus Pneumoniae In Complex With Uridine-Diphosphate-2(N-Acetylglucosaminyl) Butyric Acid, (2R)-2-(Phosphonooxy)Propanoic Acid And Magnesium.
Organism: Streptococcus pneumoniae serotype 4 (strain atcc baa-334 / tigr4)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-08-02 Classification: TRANSFERASE Ligands: 0V5, MG, EPU |
 |
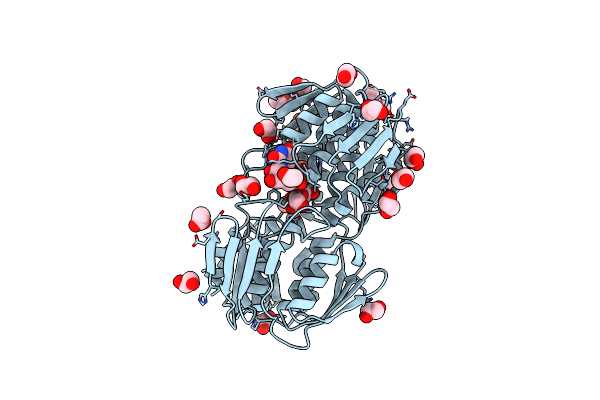
Crystal Structure Of Udp-N-Acetylglucosamine 1-Carboxyvinyltransferase (Udp-N-Acetylglucosamine Enolpyruvyl Transferase, Ept) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-07-15 Classification: TRANSFERASE Ligands: EPU, MG |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2012-03-07 Classification: TRANSFERASE Ligands: PGE, PEG, EDO, ACT, EPU |
 |
Crystal Structure Of Enolpyruvyl-Udp-Glcnac Synthase (Mura):Udp-N-Acetylmuramic Acid:Phosphite From Escherichia Coli
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-11-24 Classification: TRANSFERASE Ligands: PO3, EPU |
 |
Crystal Structure Of Udp-N-Acetylglucosamine 1-Carboxyvinyltransferase From Aquifex Aeolicus Vf5
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2007-10-16 Classification: TRANSFERASE Ligands: PO4, EPU |
 |
Crystal Structure Of Udp-N-Acetylenolpyruvylglucosamine Reductase (Murb) From Thermus Caldophilus
Organism: Thermus caldophilus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-12-19 Classification: OXIDOREDUCTASE Ligands: FAD, EPU |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-11-09 Classification: TRANSFERASE Ligands: PO4, EPU, GOL |
 |
Crystal Structure Of Udp-N-Acetylmuramic Acid:L-Alanine Ligase (Murc) From Haemophilus Influenzae
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2003-07-15 Classification: LIGASE Ligands: MG, EPU |
 |
Murb Mutant With Ser 229 Replaced By Ala, Complex With Enolpyruvyl-Udp-N-Acetylglucosamine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1997-04-01 Classification: OXIDOREDUCTASE Ligands: FAD, EPU |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1997-04-01 Classification: OXIDOREDUCTASE Ligands: FAD, EPU |

