Search Count: 8
All
Selected
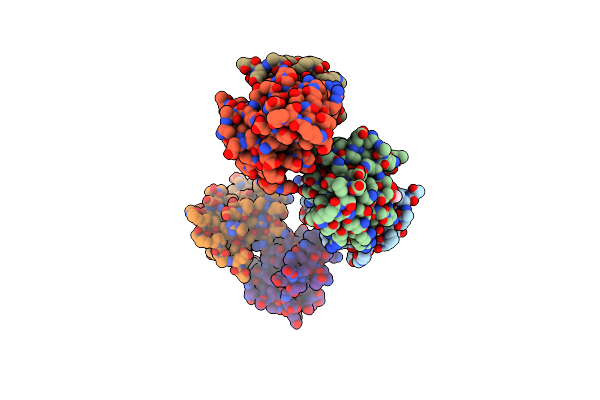 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION, EPR Resolution:1.64 Å Release Date: 2013-07-31 Classification: CHAPERONE Ligands: GOL |
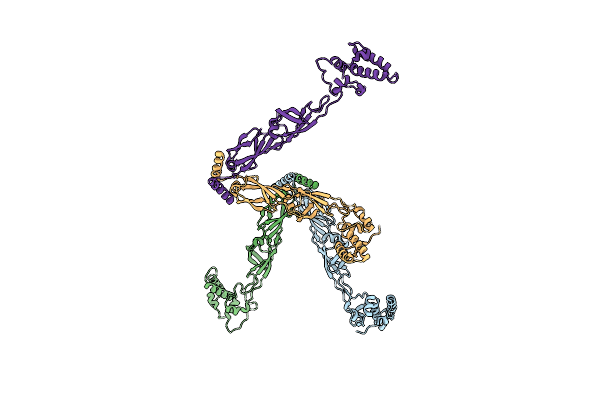 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION, EPR Resolution:2.90 Å Release Date: 2013-07-31 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: SOLUTION NMR, EPR Release Date: 2010-06-16 Classification: LIPID BINDING PROTEIN |
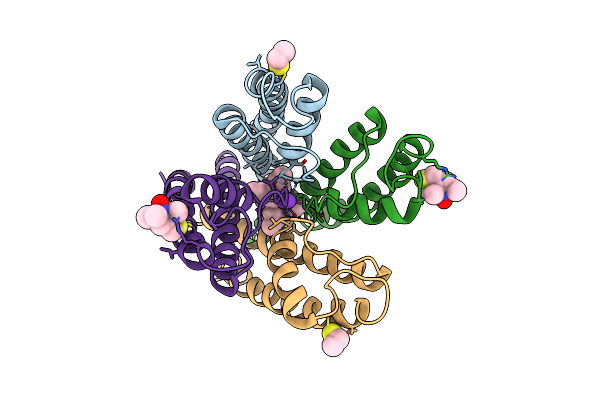 |
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION, EPR Resolution:3.56 Å Release Date: 2010-02-09 Classification: MEMBRANE PROTEIN Ligands: K, MTN, TBA |
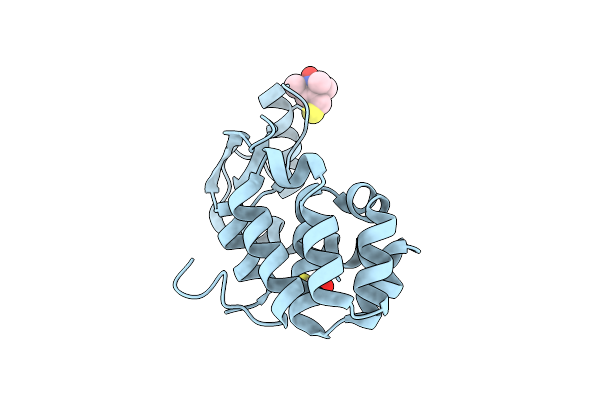 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION, EPR Resolution:1.40 Å Release Date: 2007-06-26 Classification: HYDROLASE Ligands: MTN, BME |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION, EPR Resolution:2.10 Å Release Date: 2007-06-26 Classification: HYDROLASE Ligands: MTN, HED |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION, EPR Resolution:1.80 Å Release Date: 2007-06-12 Classification: HYDROLASE Ligands: MTN, BME |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION, EPR Resolution:1.70 Å Release Date: 2006-08-15 Classification: HYDROLASE Ligands: MTN, AZI, CL, HED |

