Search Count: 2,449
All
Selected
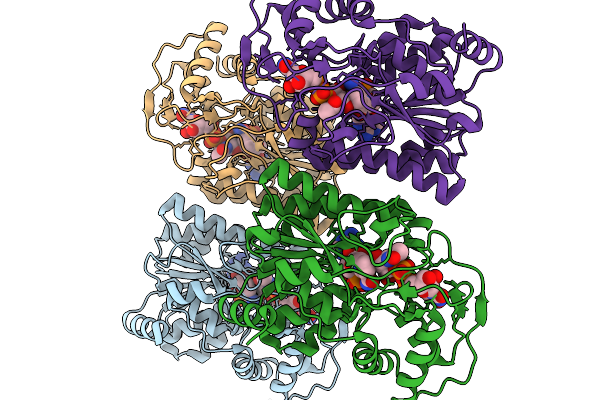 |
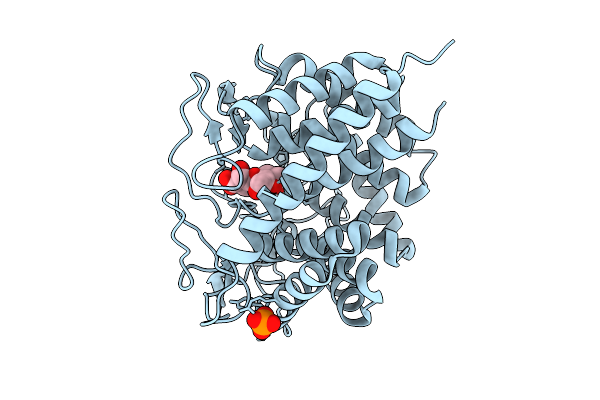
Crystal Structure Of Capsular Polysaccharide Biosynthesis Protein From Bordetella Pertussis In Complex With Nad And Uridine-Diphosphate-N-Acetylgalactosamine (Cocrystallization)
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-02-04 Classification: ISOMERASE Ligands: CL, NAD, UD2 |
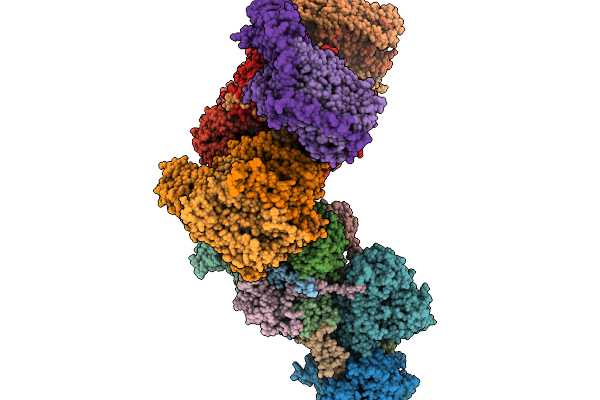 |
Organism: Paracoccus denitrificans pd1222
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: ELECTRON TRANSPORT Ligands: SF4, NA, FES, FMN, DU0, U10, ZN, CDL, HEM, CA, HEC, HEA, CU, MN, CUA |
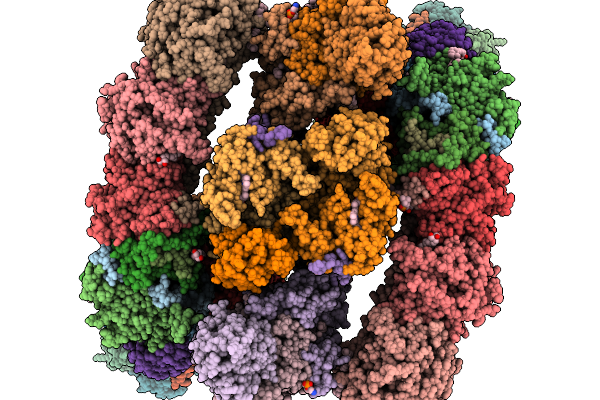 |
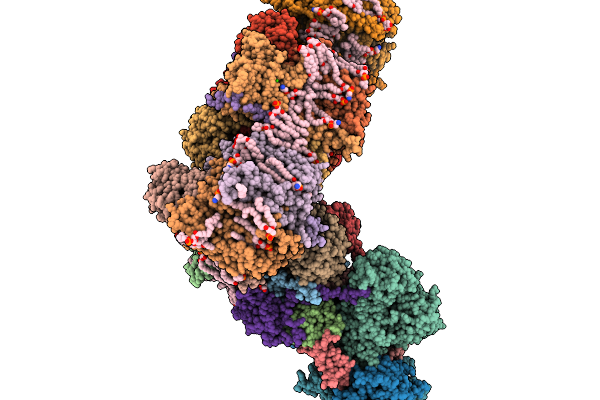
Respiratory Supercomplex Ci2-Ciii2-Civ2 (Megacomplex) From Alphaproteobacterium
|
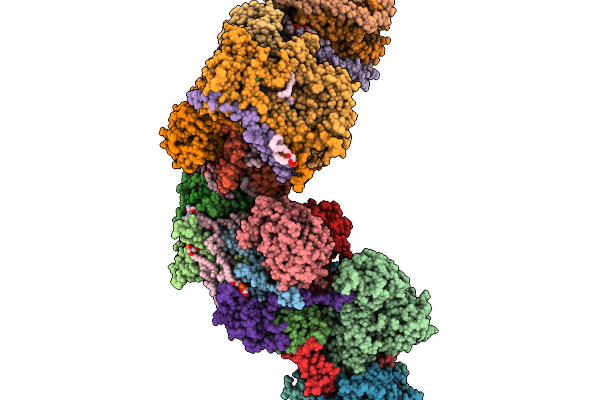 |
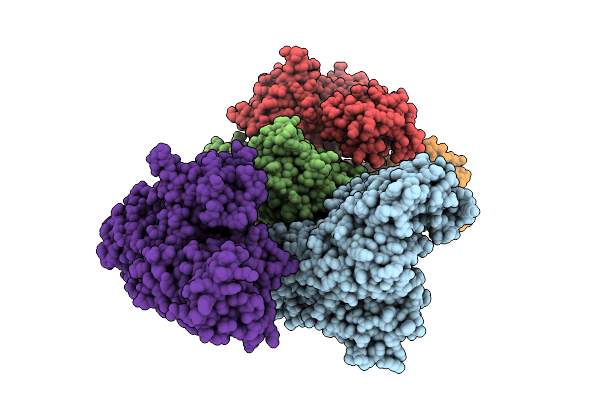
Respiratory Supercomplex Ci1-Ciii2-Civ1 (Respirasome) From Alphaproteobacterium
|
 |
 |
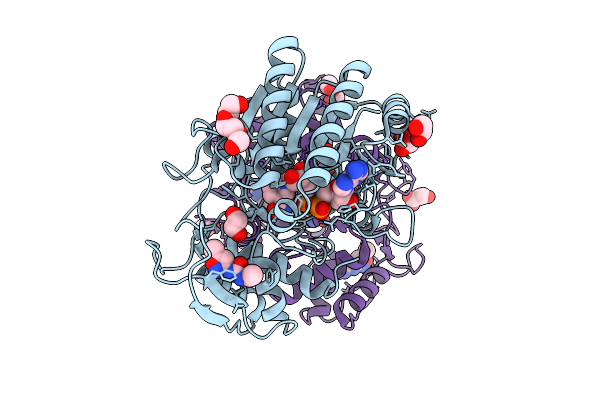
Crystal Structure Of A Wild-Type Tagose Isomerase (Tst4Ease Wt) From Thermotogota Bacterium
Organism: Thermotogota bacterium
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-12-24 Classification: ISOMERASE |
 |
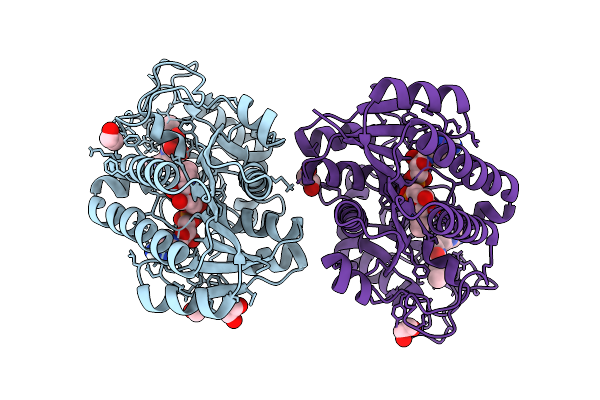
Structure Of Udp-Galactose-4-Epimerase (Gale) Bound To Fragment From Diamond Xchem Experiment.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-12-10 Classification: CARBOHYDRATE Ligands: NAD, JGA, MLI, PGE, EDO, CL |
 |
Structure Of Human Udp-Galactose 4-Epimerase In Complex With Compound Wbx04
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2025-12-03 Classification: ISOMERASE Ligands: EDO, NAD, A1IU2, MLT |
 |
Structure Of Human Udp-Galactose 4-Epimerase In Complex With Compound Wbx09
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2025-12-03 Classification: ISOMERASE Ligands: EDO, NAD, A1IU5 |
 |
Structure Of Human Udp-Galactose 4-Epimerase In Complex With Covalent Compound Wbc10
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2025-12-03 Classification: ISOMERASE Ligands: NAI, A1IU6 |
 |
Crystal Structure Of Tagatose 4-Epimerase With One Glycerol From Thermoprotei Archaeon
Organism: Thermoprotei archaeon
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2025-11-05 Classification: ISOMERASE Ligands: EDO, PEG, 1PG, NI, GOL |
 |
Organism: Thermoprotei archaeon
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-11-05 Classification: ISOMERASE Ligands: EDO, PEG, 1PG |
 |
Crystal Structure Of Apo-Form Artificial Metalloprotein Incorporating A Tp Ligand
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2025-10-15 Classification: METAL BINDING PROTEIN |
 |
Crystal Structure Of Cu-Bound Artificial Metalloprotein Incorporating A Tp Ligand
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2025-10-15 Classification: METAL BINDING PROTEIN Ligands: CU, NA, SO4 |
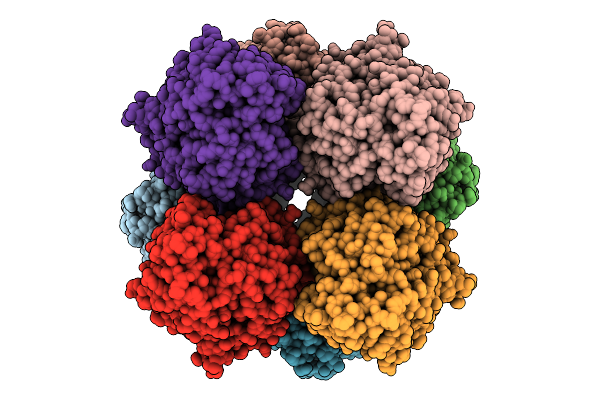 |
Structure Of Lara-Like Nickel-Pincer Nucleotide Cofactor-Utilizing Enzyme With A Single Catalytic Histidine Residue From Streptococcus Plurextorum
Organism: Streptococcus plurextorum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: ISOMERASE Ligands: NI, 4EY |
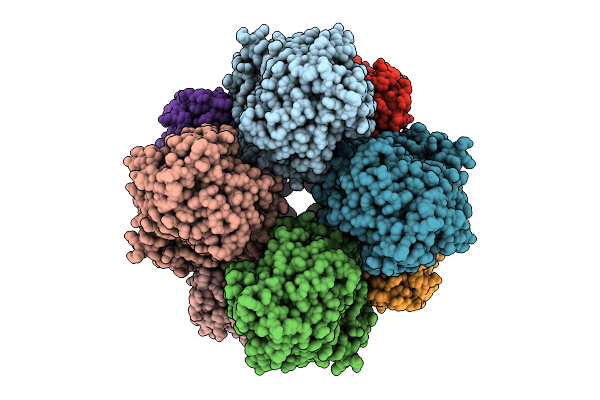 |
Structures Of Lara-Like Nickel-Pincer Nucleotide Cofactor-Utilizing Enzyme With A Single Catalytic Histidine Residue From Blautia Wexlerae
Organism: Blautia wexlerae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: ISOMERASE |
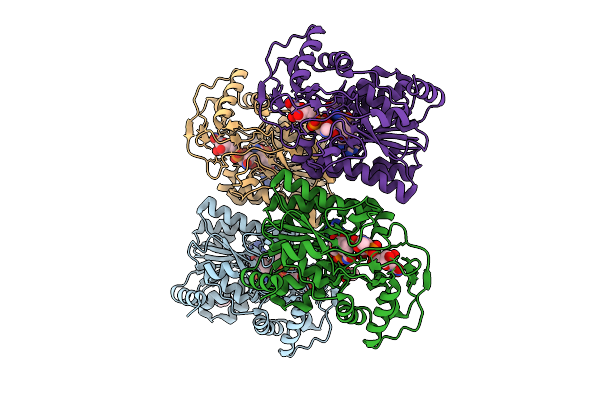 |
Crystal Structure Of Capsular Polysaccharide Biosynthesis Protein From Bordetella Pertussis In Complex With Nad And Uridine-Diphosphate-N-Acetylgalactosamine
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-08-06 Classification: ISOMERASE Ligands: CL, NAD, UD2, ACT |
 |
Rhodothermus Marines Cellobiose 2-Epimerase Rmce In Complex With Mannobiose
Organism: Rhodothermus marinus jcm 9785
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-23 Classification: ISOMERASE Ligands: CL, PO4 |
 |
Rhodothermus Marines Cellobiose 2-Epimerase Rmce In Complex With Mannobiitol
Organism: Rhodothermus marinus jcm 9785
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-23 Classification: ISOMERASE Ligands: MTL, BMA, CL, PO4 |
 |
Crystal Structures Of A Cyanobacterial Dap Epimerase Bound To Either D,L-Azidap Or D,L-Alpha-Methyl Dap
Organism: Anabaena sp. ybs01
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2025-07-16 Classification: ANTIBIOTIC Ligands: ZDR |

