Search Count: 184
All
Selected
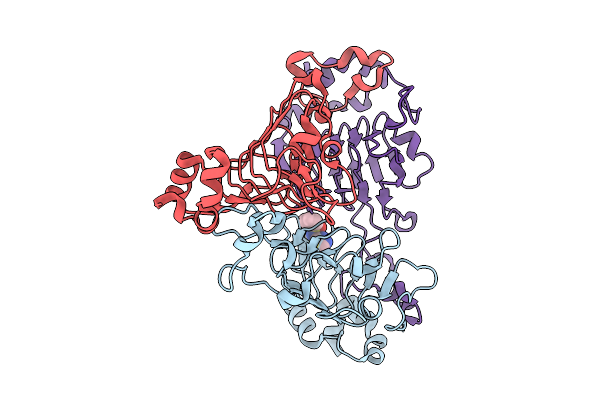 |
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000000438614
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: W6G |
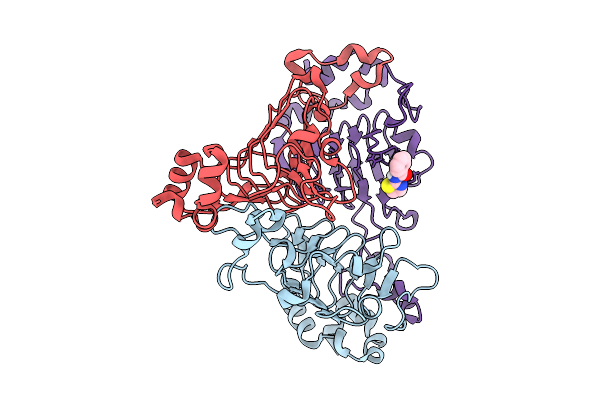 |
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000000039994
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: W6V |
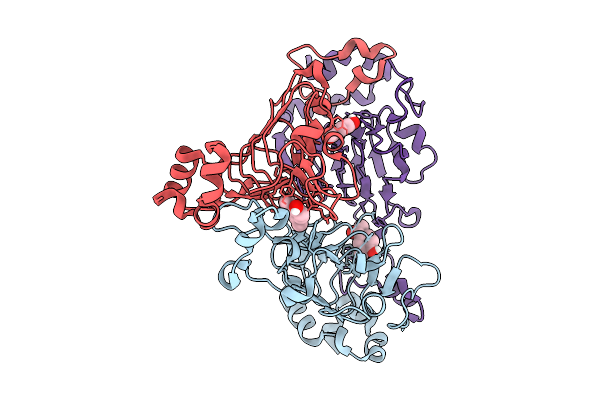 |
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000000164888
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: YTP |
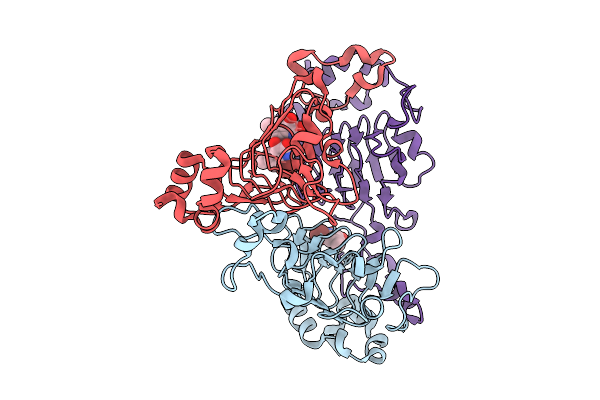 |
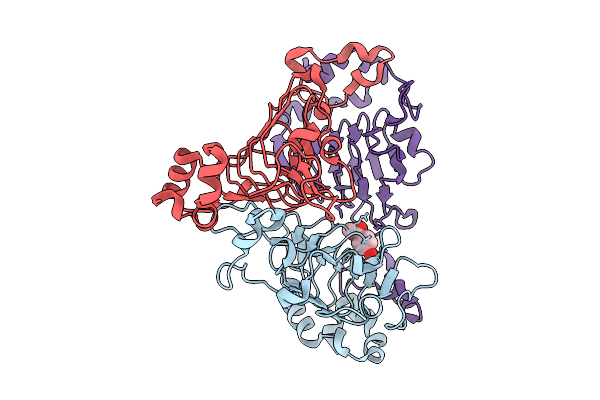
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000002977810
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: 2AK |
 |
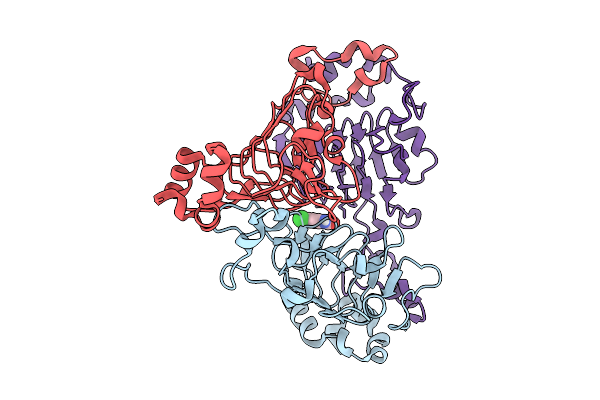
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000000001712
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: MPB |
 |
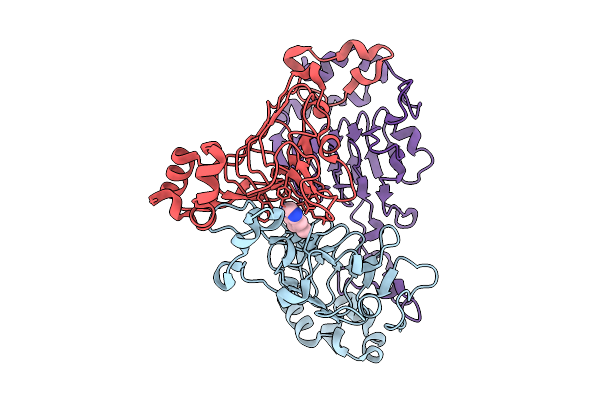
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000084843283
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: CLW |
 |
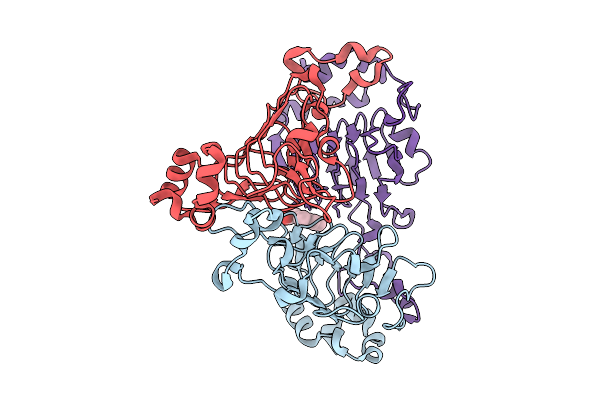
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000000388593
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: DMD |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000000873830
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: NMI |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000027858187
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: EVD |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000002561227
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: 47S |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000016889973
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: AX7 |
 |
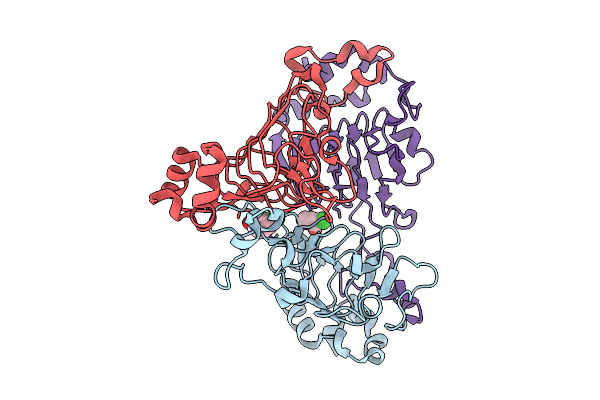
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000001389101
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: 7MU |
 |
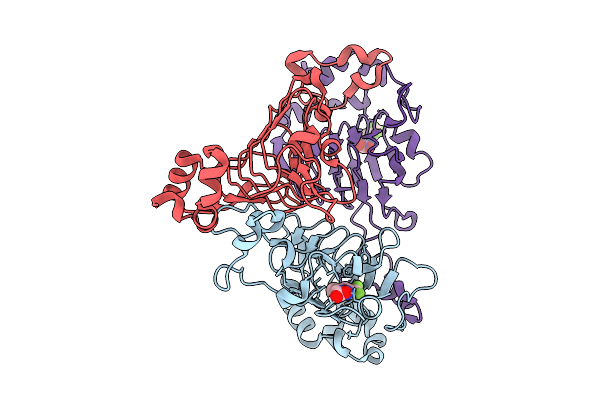
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000001683100
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: W6A |
 |
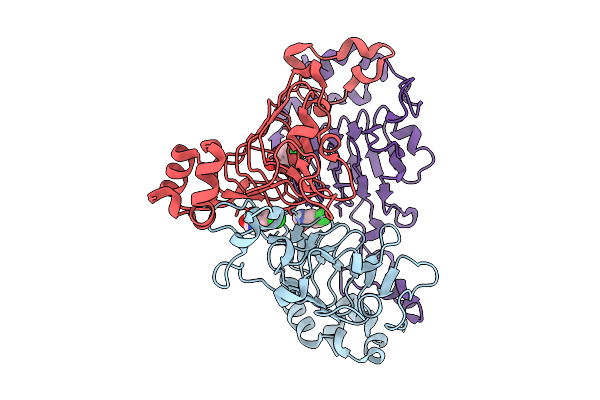
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000004976927
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: 04R |
 |
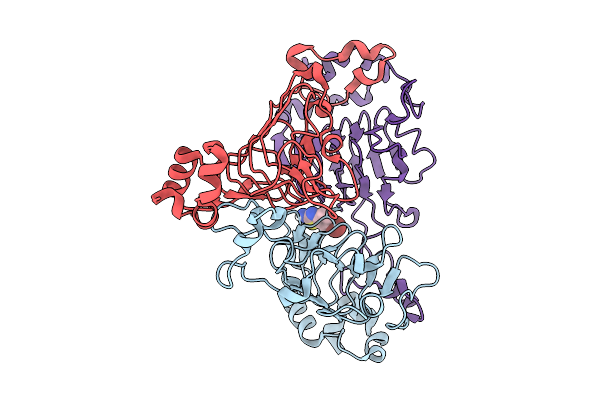
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000020269204
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: A1BUQ |
 |
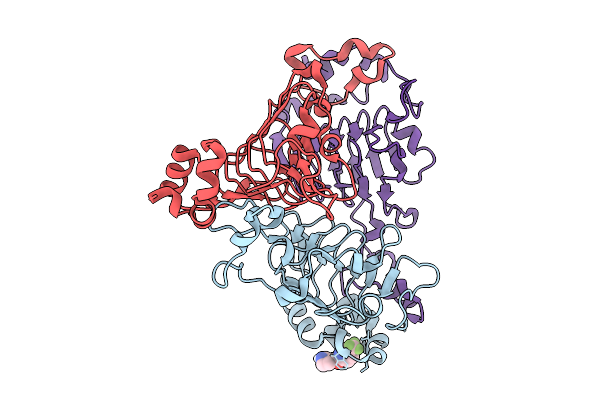
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000004774482
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: A1H0M |
 |
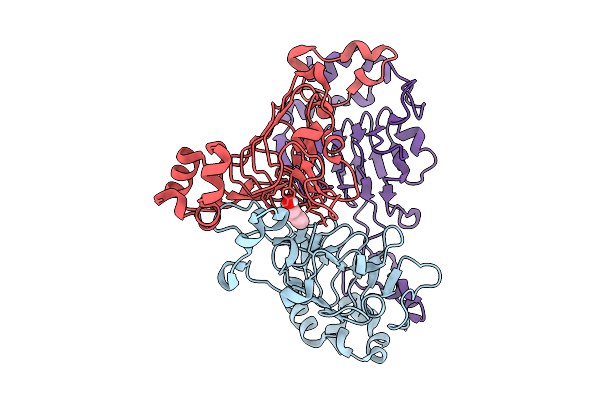
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000042783023
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: 80Q |
 |
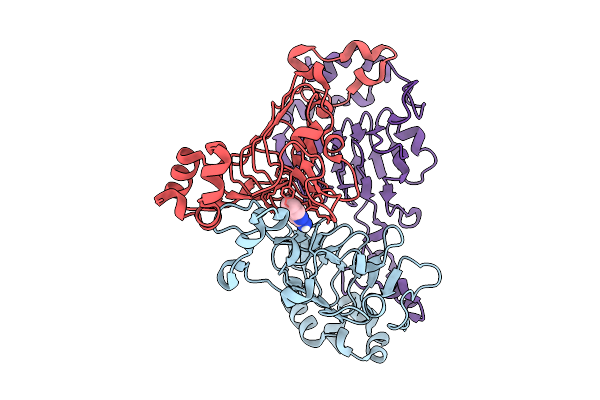
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000000388332
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: 4FA |
 |
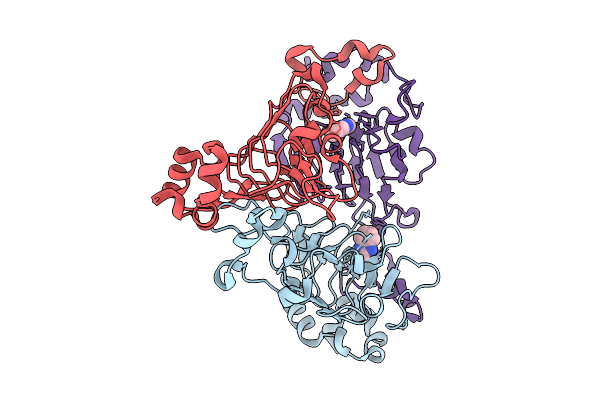
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000019074717
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: 2AQ |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Enterococcus Faecium Vatd In Complex With Zinc000000164607
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: 8P7 |

