Search Count: 14
 |
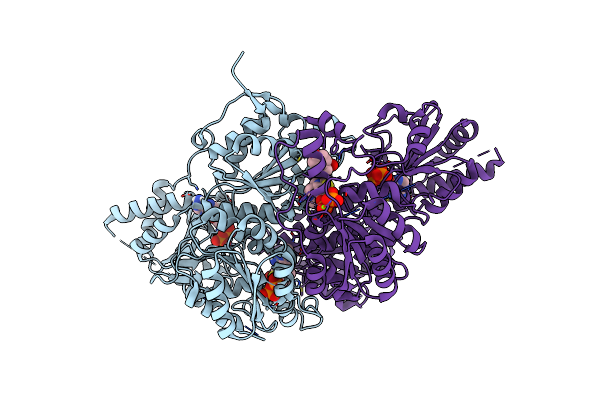
Joint Xray/Neutron Structure Of Macrophage Migration Inhibitory Factor (Mif) Bound To 4-Hydroxyphenylpyruvate At Room Temperature
Organism: Homo sapiens
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-08-27 Classification: CYTOKINE Ligands: ENO, IPA, EN1 |
 |
Organism: Brevibacillus
Method: X-RAY DIFFRACTION Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN Ligands: TPP, ADP, ENO, MG |
 |
X-Ray Structure Of Macrophage Migration Inhibitory Factor (Mif) Covalently Bound To 4-Hydroxyphenylpyruvate (Hpp)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-08-28 Classification: CYTOKINE Ligands: ENO, IPA |
 |
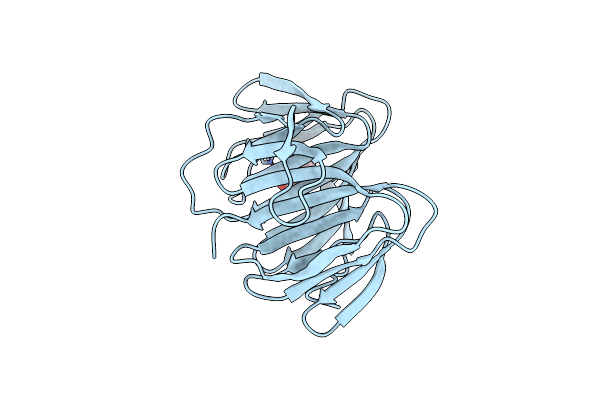
Crystal Structure Of Nadph And 4-Hydroxyphenylpyruvic Acid Bound Aerf From Microcystis Aeruginosa
Organism: Microcystis aeruginosa dianchi905
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2019-11-27 Classification: BIOSYNTHETIC PROTEIN Ligands: NDP, ENO |
 |
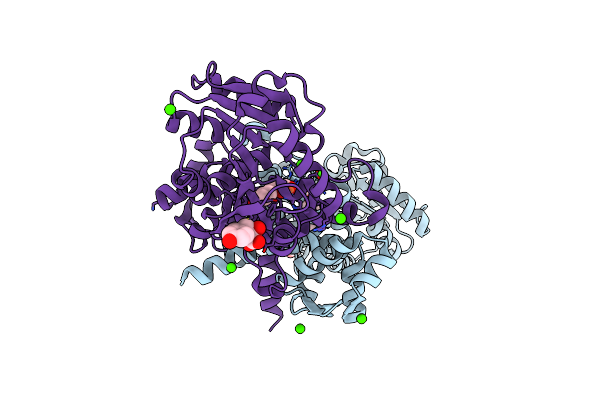
Crystal Structure Of 4-Hydroxyphenylpyruvic Acid Bound Aere From Microcystis Aeruginosa
Organism: Microcystis aeruginosa dianchi905
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-02-13 Classification: ISOMERASE Ligands: ENO, FE2 |
 |
Crystal Structure Of Methyltransferase From Streptomyces Hygroscopicus Complexed With 4-Hydroxyphenylpyruvic Acid
Organism: Streptomyces hygroscopicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-05-07 Classification: TRANSFERASE Ligands: ENO, FE, 34H, CA |
 |
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2013-04-17 Classification: TRANSPORT PROTEIN Ligands: FER, ENO, GOL |
 |
Crystal Structure Of Solute Binding Protein Of Abc Transporter From Rhodopseudomonas Palustris Haa2 In Complex With Caffeic Acid/3-(4-Hydroxy-Phenyl)Pyruvic Acid
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2012-05-30 Classification: TRANSPORT PROTEIN Ligands: ENO, DHC |
 |
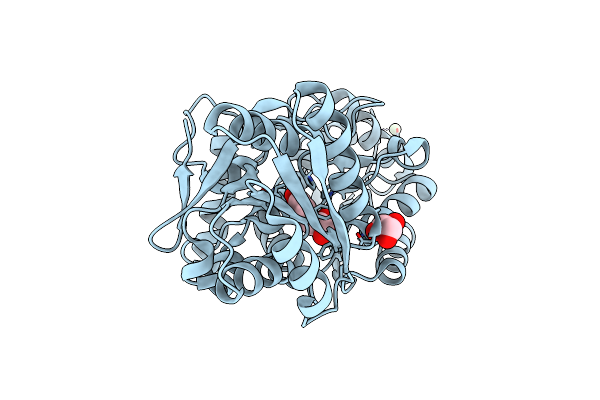
Rpd_1889 Protein, An Extracellular Ligand-Binding Receptor From Rhodopseudomonas Palustris.
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2011-11-23 Classification: TRANSPORT PROTEIN Ligands: ENO, SO4, EDO |
 |
Crystal Structure Of Extracellular Ligand-Binding Receptor From Rhodopseudomonas Palustris Haa2
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-11-23 Classification: TRANSPORT PROTEIN Ligands: ENO, PR, GOL |
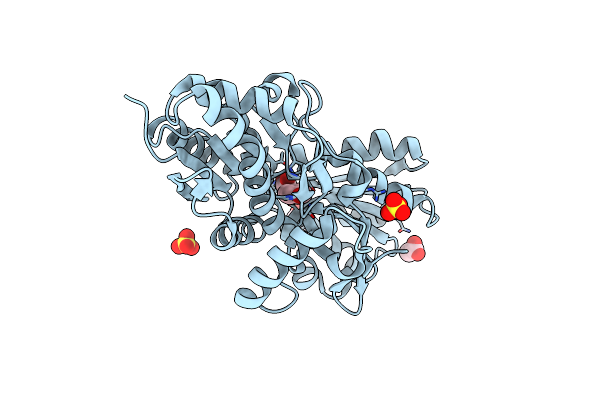 |
The Structure Of A Putative Abc-Transporter Periplasmic Component From Rhodopseudomonas Palustris
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-11-09 Classification: SIGNALING PROTEIN Ligands: SO4, ENO, EDO, ACY, CL |
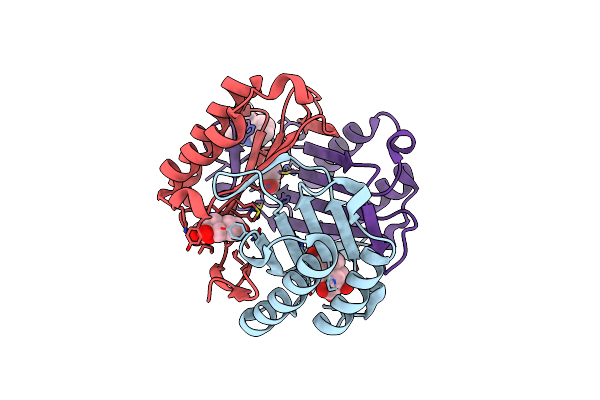 |
Ternary Complex Of Macrophage Migration Inhibitory Factor (Mif) Bound Both To 4-Hydroxyphenylpyruvate And To The Allosteric Inhibitor Av1013 (R-Stereoisomer)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2010-06-16 Classification: ISOMERASE Ligands: EN1, ENO, CL, AVR |
 |
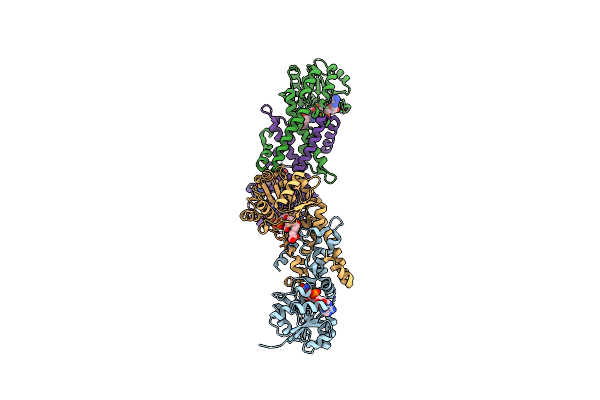
Organism: Plasmodium yoelii yoelii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-12-29 Classification: CYTOKINE Ligands: SO4, ACY, ENO |
 |
Crystal Structure Of Prephenate Dehydrogenase From A. Aeolicus With Hpp And Nadh
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2009-03-10 Classification: OXIDOREDUCTASE Ligands: NAI, ENO |

