Search Count: 615
 |
Organism: Syntrophomonas palmitatica jcm 14374, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-11 Classification: DNA BINDING PROTEIN Ligands: ZN, FE2, EPE, SO4, NI, CL |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-07-10 Classification: HYDROLASE Ligands: FE, ZN, PO4, CL |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2023-09-13 Classification: HYDROLASE Ligands: FE, ZN, SO4 |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-09-06 Classification: HYDROLASE Ligands: FE, ZN, PO4, CL |
 |
Cas1-Cas2/3 Integrase And Ihf Bound To Crispr Leader, Repeat And Foreign Dna
Organism: Pseudomonas aeruginosa pa14, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.48 Å Release Date: 2023-09-06 Classification: RECOMBINATION, DNA BINDING PROTEIN, HYDROLASE/DNA |
 |
Crystal Structure Of K74A Mutant Of Cap4 Saved Domain-Containing Receptor From Enterobacter Cloacae
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2023-05-31 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of Wild-Type Cap4 Saved Domain-Containing Receptor From Enterobacter Cloacae
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2023-05-31 Classification: HYDROLASE Ligands: CL |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-08-17 Classification: HYDROLASE Ligands: MG |
 |
Organism: Deinococcus radiodurans atcc 13939
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-07-06 Classification: DNA BINDING PROTEIN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2020-02-26 Classification: DNA BINDING PROTEIN |
 |
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.41 Å Release Date: 2019-09-25 Classification: HYDROLASE |
 |
Crystal Structure Of The Apurinic/Apyrimidinic Endonuclease Iv From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2018-04-04 Classification: DNA BINDING PROTEIN Ligands: SO4, ZN |
 |
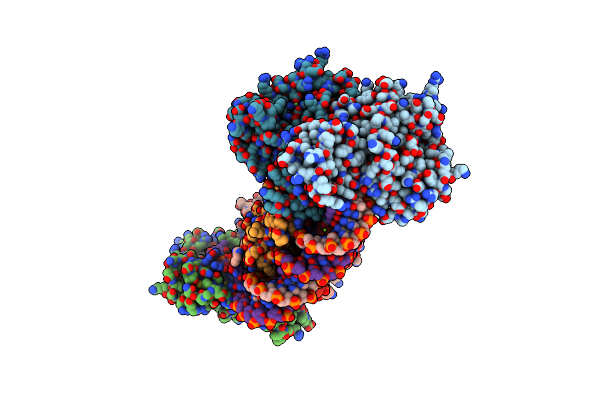
Crystal Structure The Escherichia Coli Cas1-Cas2 Complex Bound To Protospacer Dna
Organism: Escherichia coli (strain k12), Enterobacteria phage m13
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-10-28 Classification: Hydrolase/DNA |
 |
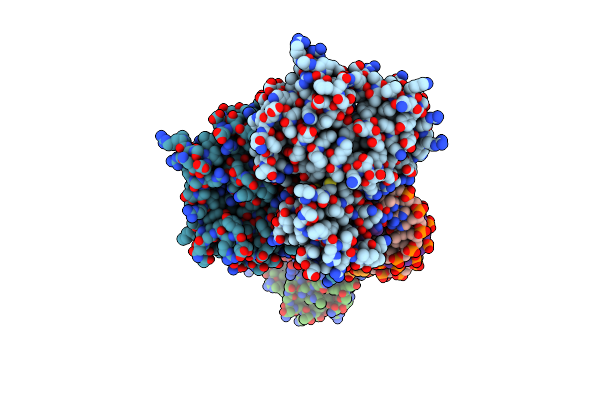
Crystal Structure The Escherichia Coli Cas1-Cas2 Complex Bound To Protospacer Dna And Mg
Organism: Escherichia coli (strain k12), Enterobacteria phage m13
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2015-10-28 Classification: Hydrolase/DNA Ligands: MG |
 |
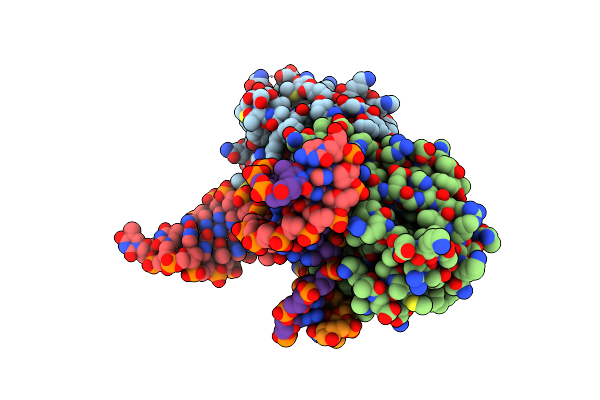
Crystal Structure The Escherichia Coli Cas1-Cas2 Complex Bound To Protospacer Dna With Splayed Ends
Organism: Escherichia coli (strain k12), Enterobacteria phage m13
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2015-10-28 Classification: Hydrolase/DNA |
 |
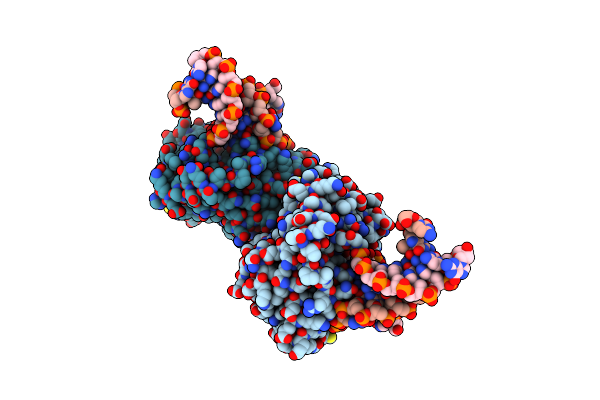
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.75 Å Release Date: 2013-09-04 Classification: HYDROLASE/DNA |
 |
Structure Of E. Coli Nfo(Endo Iv)-H69A Mutant Bound To A Cleaved Dna Duplex Containing A Alphada:T Basepair
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-07-31 Classification: HYDROLASE/DNA Ligands: ZN, PEG |
 |
High Resolution Crystal Structure Of Dna Apurinic/Apyrimidinic (Ap) Endonuclease Iv Nfo From Thermatoga Maritima
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:0.92 Å Release Date: 2013-01-23 Classification: HYDROLASE Ligands: ZN, MN, MG, TRS, EDO, SO4 |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2012-11-14 Classification: HYDROLASE Ligands: MG, GOL |

