Search Count: 2,676
 |
Organism: Geobacter sulfurreducens pca
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2026-01-21 Classification: ELECTRON TRANSPORT Ligands: GOL, SO4 |
 |
Organism: Geobacter sulfurreducens pca
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2026-01-21 Classification: ELECTRON TRANSPORT Ligands: SO4, GOL |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Cyclobutrifluram-Bound State
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: FAD, A1EKU, FES, SF4, F3S, PEE |
 |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: ELECTRON TRANSPORT Ligands: HEO, HEM, CU, ZN |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2025-12-24 Classification: ELECTRON TRANSPORT Ligands: CU, TB |
 |
Crystal Structure Of C43S Terpredoxin, A [2Fe-2S] Ferredoxin From Pseudomonas Sp
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-12-24 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
 |
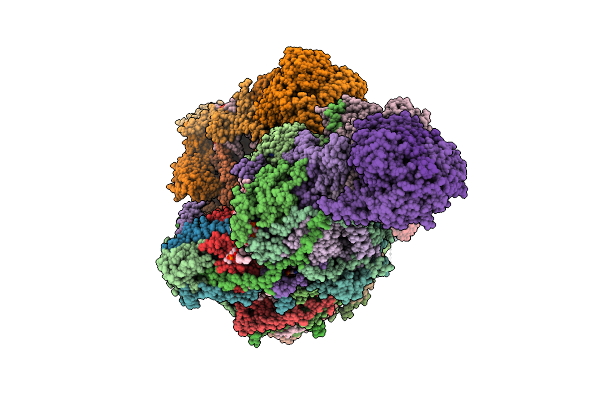
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: CDL, PEE, NDP, ZMP, ADP, 3PE, PC1, PLX, XEW, SF4, FES, MG, ZN, FMN |
 |
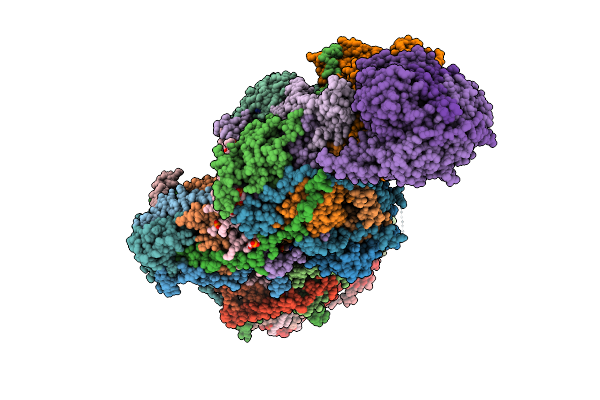
Organism: Paracoccus denitrificans pd1222
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: ELECTRON TRANSPORT Ligands: PC1, U10, HEM, CA, HEC, 3PE, DU0, FES, 3PH, CDL, HEA, CU, MN, CUA, ZN |
 |
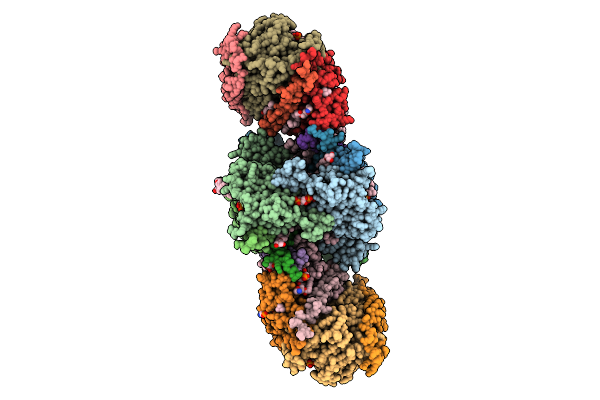
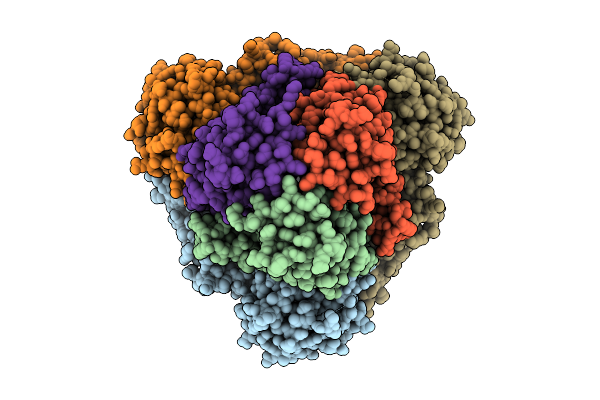
Cryo-Em Structure Of Spinacia Oleracea Cytochrome B6F Complex With Bound Plastocyanin
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Resolution:3.18 Å Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: HEM, HEC, UMQ, PL9, CLA, FES, SQD, BCR, CU |
 |
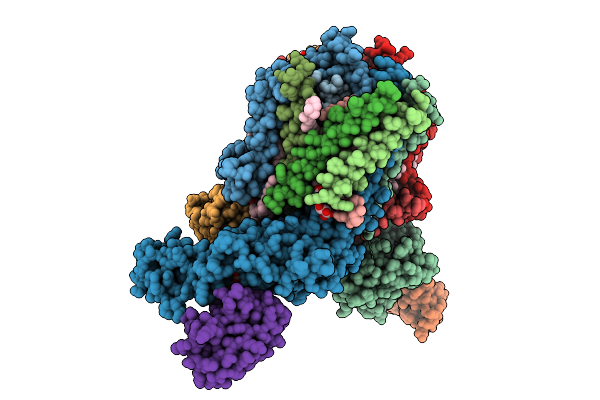
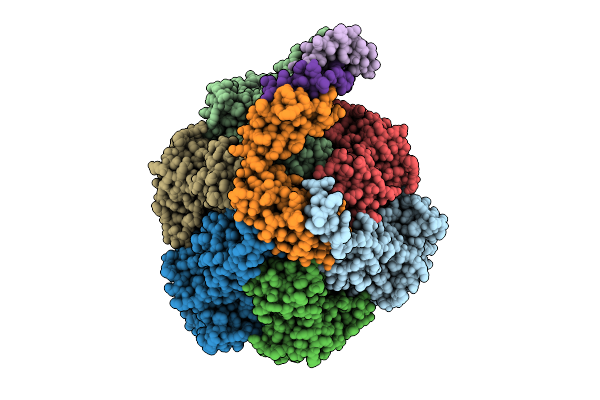
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: FE2, FES, HCI |
 |
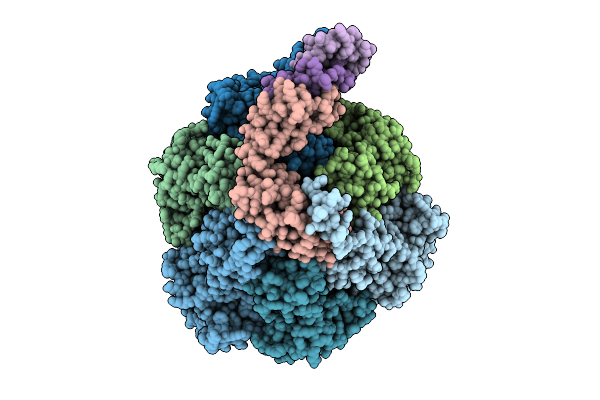
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, PO4 |
 |
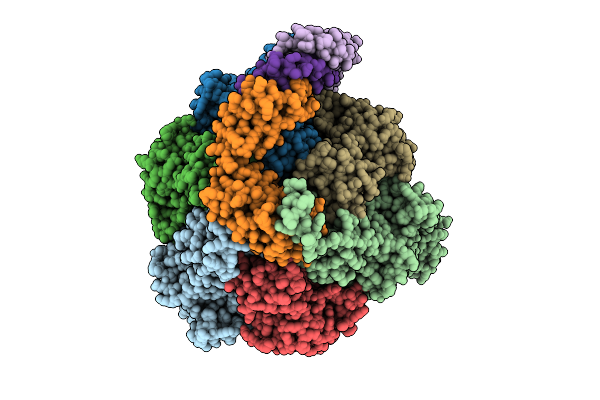
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP, ZN |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: ZN |

