Search Count: 1,033
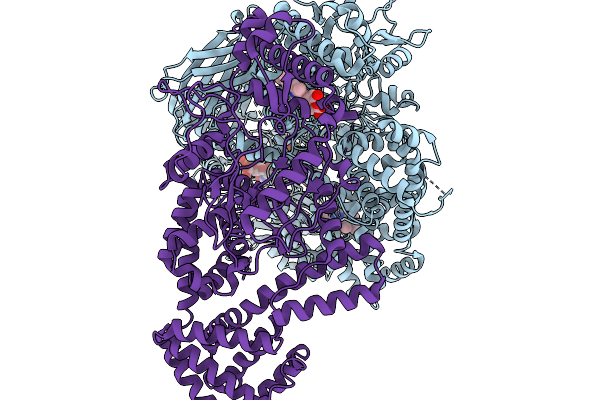 |
Organism: Synechococcus sp. pcc 7335
Method: ELECTRON MICROSCOPY Resolution:3.19 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: HEM, FAD, FMN |
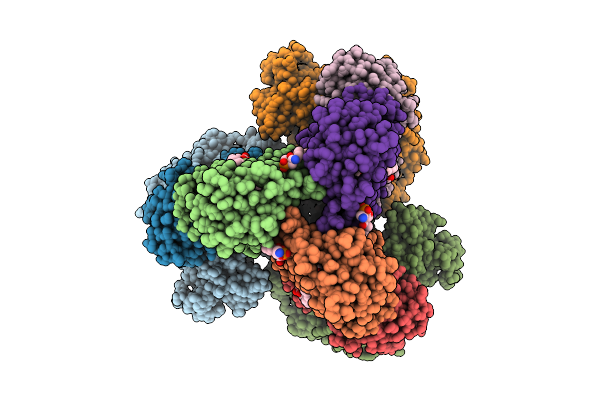 |
Cryo-Em Structure Of The Apo-Form Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SIN, SF4, FES, F3S, HEM, LMT, PGV, PEV |
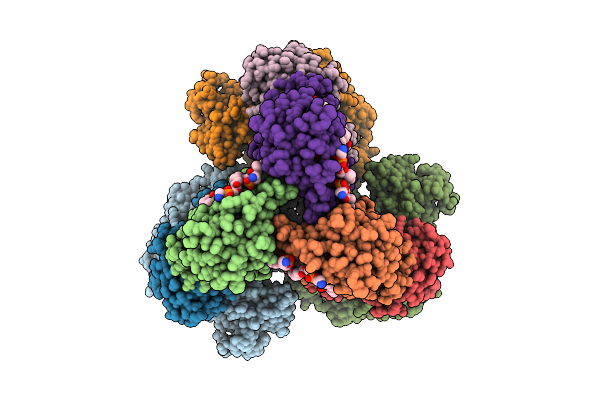 |
Cryo-Em Structure Of The Lipid-Bound Succiante Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SIN, SF4, FES, F3S, HEM, LMT, PGV, PEV |
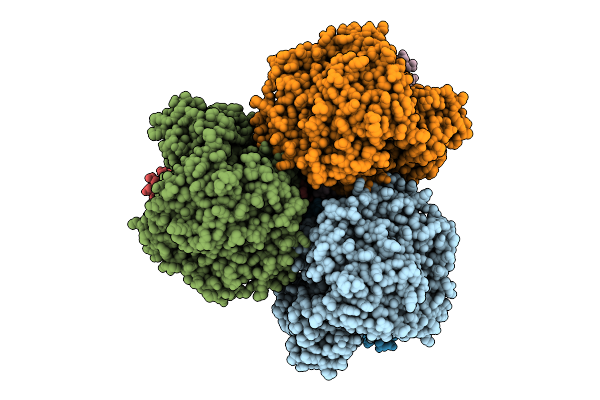 |
Cryo-Em Structure Of The Mk7-Bound Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SF4, FES, F3S, HEM, LMT, MQ7 |
 |
Cryo-Em Structure Of The Mk4-Bound Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SF4, FES, F3S, HEM, LMT, 1L3 |
 |
Crystal Structure Of Domains I And Ii From The Outer Membrane Cytochrome Mtrc
Organism: Shewanella oneidensis mr-1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-08-13 Classification: ELECTRON TRANSPORT Ligands: HEC, MG, EDO |
 |
Crystal Structure Of Reduced Wild Type Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2025-07-09 Classification: OXIDOREDUCTASE Ligands: CU, HEC, SO4 |
 |
Crystal Structure Of Nitric Oxide Treated F295L Mutant Of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2025-07-09 Classification: OXIDOREDUCTASE Ligands: CU, HEC, NO |
 |
Crystal Structure Of Reduced F295L Mutant Of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2025-07-09 Classification: OXIDOREDUCTASE Ligands: CU, HEC |
 |
Crystal Structure Of Nitric Oxide-Treated Q262N Mutant Of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2025-07-09 Classification: OXIDOREDUCTASE Ligands: CU, HEC |
 |
Crystal Structure Of As-Isolated F295L Mutant Of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: CU, HEC |
 |
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:0.92 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: FES |
 |
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: FES, SO4 |
 |
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: FES |
 |
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: FES |
 |
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: FES |
 |
Apo Fefe Hydrogenase From Desulfovibrio Desulfuricans Labelled With Cyanophenylalanine
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: SF4, CL, LI |
 |
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-05-28 Classification: ELECTRON TRANSPORT Ligands: HEC, SO4, ACY, GOL, PEG |
 |
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Resolution:3.68 Å Release Date: 2025-05-07 Classification: OXIDOREDUCTASE Ligands: SF4, PMR, CU |
 |
Cryoem Structure Of Bchn-Bchb Bound To Pchlide From The Dpor Under Turnover Complex Dataset
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Resolution:3.29 Å Release Date: 2025-05-07 Classification: OXIDOREDUCTASE Ligands: SF4, PMR, CU |

