Search Count: 14
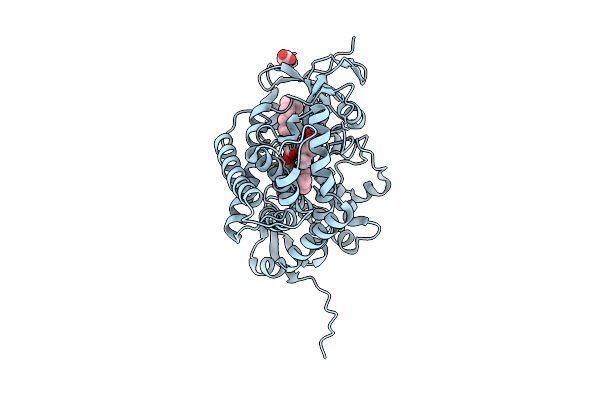 |
Cytochrome P450 Decarboxylase From Staphylococcus Aureus (Olet_Sa) With Elaidic Acid And Acetate Bound
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: HEM, ELA, ACT |
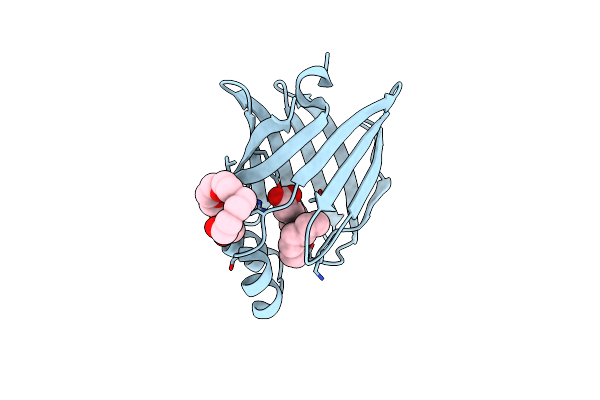 |
The 0.86 Angstrom X-Ray Structure Of The Human Heart Fatty Acid-Binding Protein Complexed With Elaidic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-07-20 Classification: LIPID BINDING PROTEIN Ligands: ELA, P6G |
 |
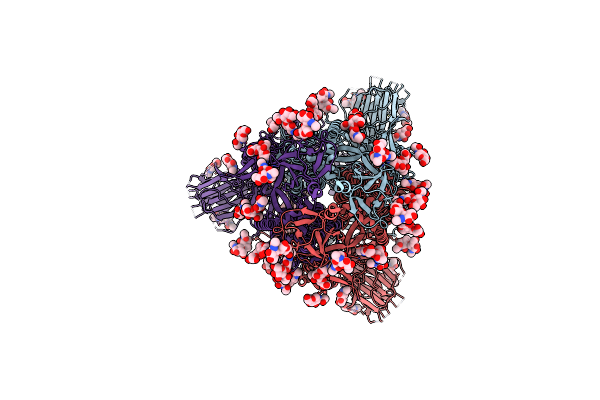
Cryo-Em Structure Of The Sars-Cov-2 Furin Site Mutant S-Trimer From A Subunit Vaccine Candidate
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-24 Classification: VIRAL PROTEIN Ligands: NAG, ELA, VCG |
 |
Cryo-Em Structure Of The Sars-Cov-2 Wild-Type S-Trimer From A Subunit Vaccine Candidate
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-24 Classification: VIRAL PROTEIN Ligands: NAG, ELA |
 |
Bacteriophage Phikz Gp163.1 Paar Repeat Protein In Complex With The C-Terminal Part Of The T4 Gp5 Beta-Helical Domain
Organism: Enterobacteria phage t4, Pseudomonas phage phikz
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: viral protein, hydrolase Ligands: MG, PLM, STE, ELA, ZN |
 |
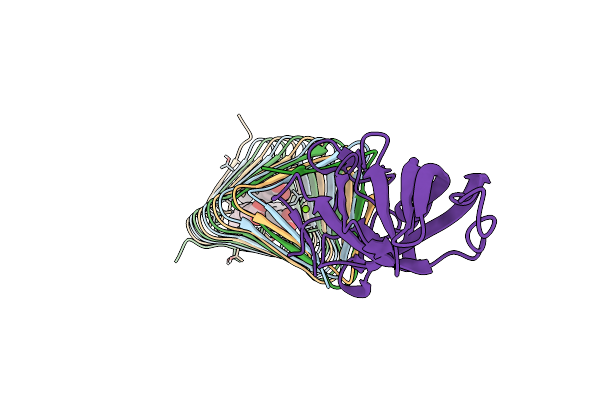
Bacteriophage Phikz Gp163.1 Paar Repeat Protein In Complex With A T4 Gp5 Beta-Helix Fragment Modified To Mimic The Phikz Central Spike Gp164
Organism: Enterobacteria phage t4, Pseudomonas phage phikz
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: VIRAL PROTEIN Ligands: MG, STE, ELA, EDO, PLM, ZN |
 |
Photorhabdus Virulence Cassette (Pvc) Paar Repeat Protein Pvc10 In Complex With A T4 Gp5 Beta-Helix Fragment Modified To Mimic Pvc8, The Central Spike Protein Of Pvc
Organism: Enterobacteria phage t4, Photorhabdus luminescens subsp. laumondii (strain dsm 15139 / cip 105565 / tt01)
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: VIRAL PROTEIN Ligands: MG, STE, ELA, PLM |
 |
Chimera Of Bacteriophage Obp Gp146 Central Spike Protein And A T4 Gp5 Beta-Helix Fragment
Organism: Enterobacteria phage t4, Pseudomonas phage obp
Method: X-RAY DIFFRACTION Release Date: 2020-05-27 Classification: VIRAL PROTEIN Ligands: FE2, STE, ELA, PLM, MG |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Release Date: 2015-02-18 Classification: HYDROLASE Ligands: ELA, MG, STE, PLM |
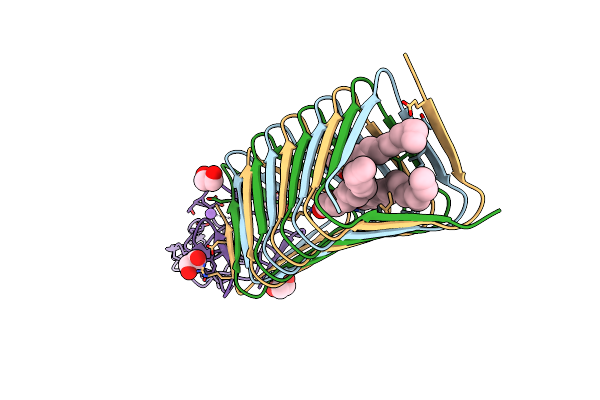 |
Enterobacteria Phage T4 Gp5.4 Paar Repeat Protein In Complex With T4 Gp5 Beta-Helix Fragment
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Release Date: 2014-05-21 Classification: HYDROLASE/PROTEIN BINDING Ligands: MG, ELA, EDO, STE, PLM, FE, NA |
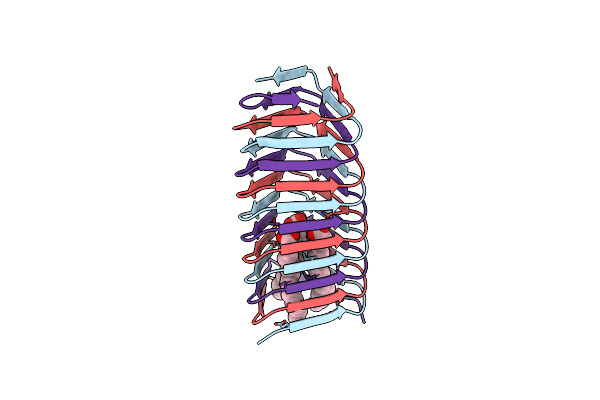 |
High Resolution Structure Of A C-Terminal Fragment Of The T4 Phage Gp5 Beta-Helix
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Release Date: 2014-03-12 Classification: HYDROLASE Ligands: MG, ELA, PLM, STE |
 |
Vca0105 Paar-Repeat Protein From Vibrio Cholerae In Complex With A Vgrg-Like Beta-Helix That Is Based On A Fragment Of T4 Gp5
Organism: Enterobacteria phage t4, Vibrio cholerae o1 biovar eltor
Method: X-RAY DIFFRACTION Release Date: 2013-08-14 Classification: HYDROLASE/PROTEIN BINDING Ligands: MG, PLM, STE, ELA, ZN |
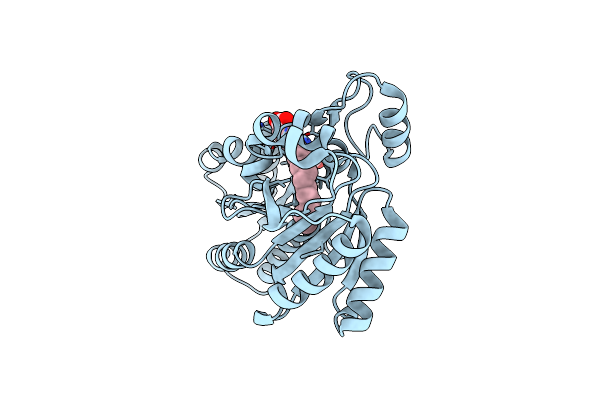 |
Crystal Structure Of Fatty Acid Binding Degv Family Protein Sag1342 From Streptococcus Agalactiae
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Release Date: 2010-03-02 Classification: structure genomics, unknown function Ligands: ELA, GOL |
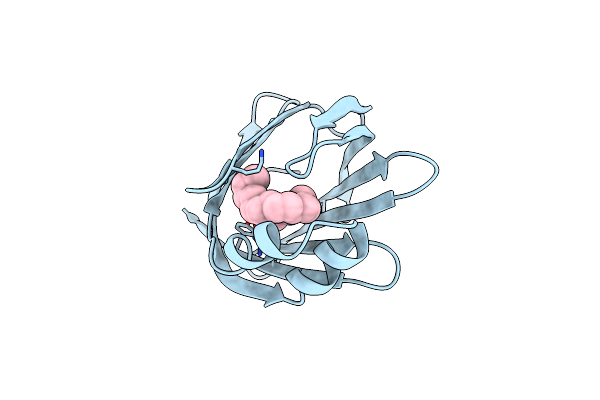 |
1.4 Angstroms Structural Studies On Human Muscle Fatty Acid Binding Protein: Binding Interactions With Three Saturated And Unsaturated C18 Fatty Acids
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 1995-05-08 Classification: LIPID BINDING PROTEIN Ligands: ELA |

