Search Count: 21
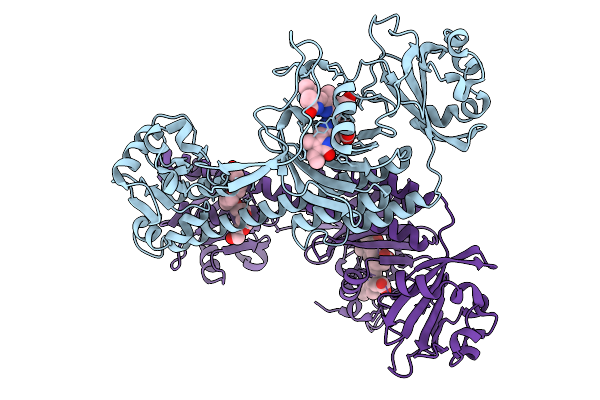 |
Photoactivation In Bacteriophytochrome, High Resolution Cryo Structure In The Dark.
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, P33 |
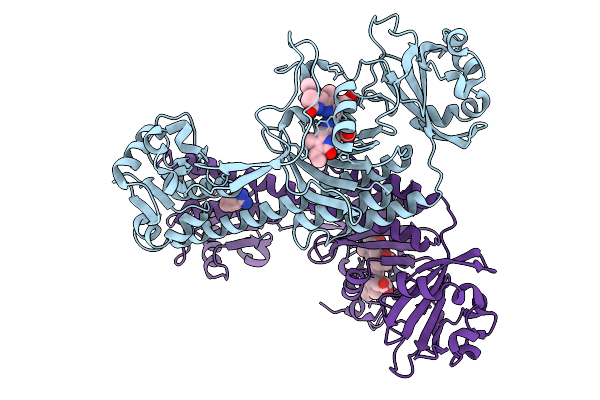 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 100 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, BEN |
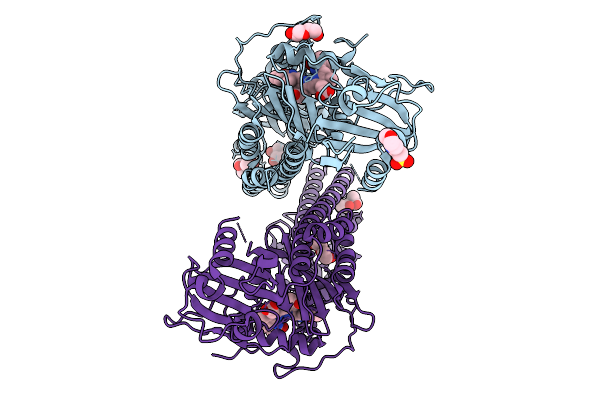 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Cyano-Phenylalanine Mutant Ocnf205 Of The Bathy Phytochrome Agp2 From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, MPD, PEG, MES, CL |
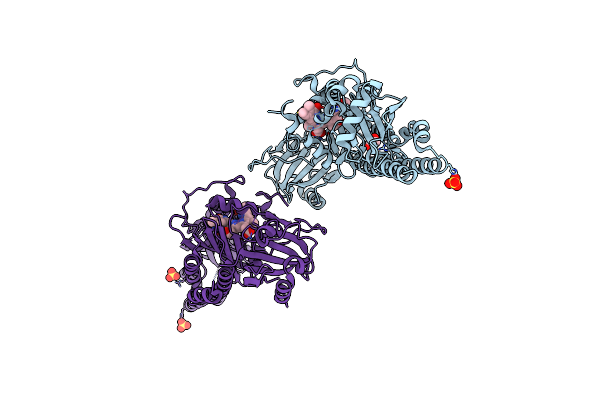 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
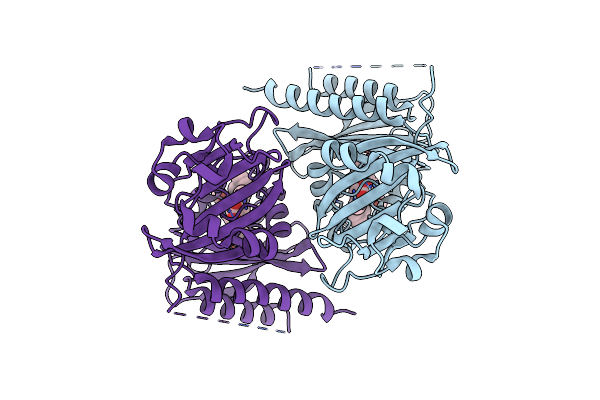 |
Unusual Structure Of A Bacteriophytochrome Fragment Derived From Full Length Sabphp2
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5 |
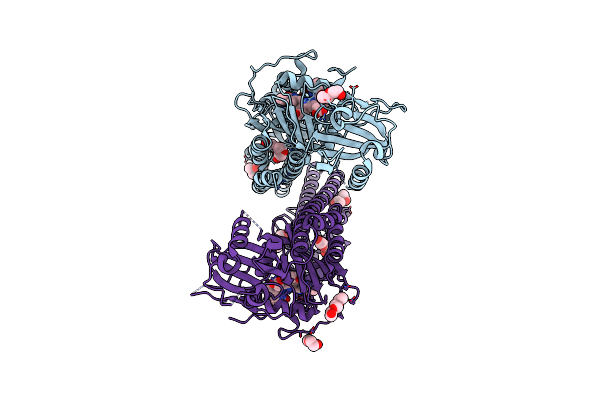 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Cyano-Phenylalanine Mutant Ocnf165 Of The Bathy Phytochrome Agp2 From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-11-27 Classification: SIGNALING PROTEIN Ligands: P6G, MPD, PEG, EL5, CL |
 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Cyano-Phenylalanine Mutant Ocnf192 Of The Bathy Phytochrome Agp2 From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2024-11-27 Classification: SIGNALING PROTEIN Ligands: PEG, MPD, EL5, CL |
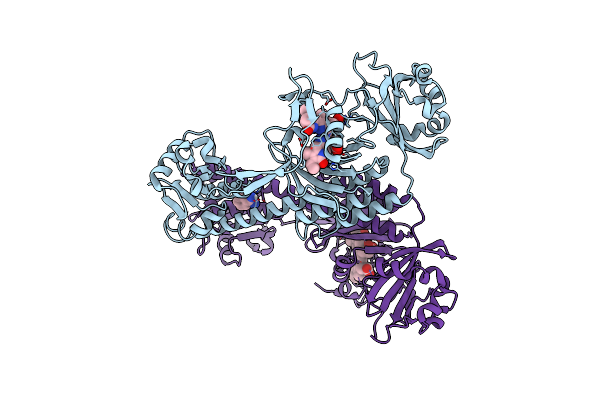 |
Organism: Stigmatella aurantiaca dw4/3-1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-10-09 Classification: SIGNALING PROTEIN Ligands: EL5, BEN |
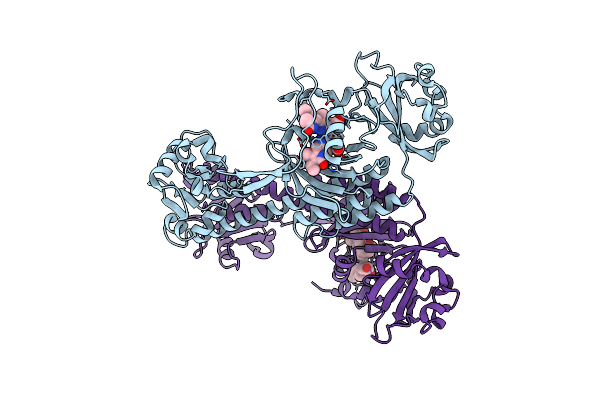 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-10-09 Classification: SIGNALING PROTEIN Ligands: EL5 |
 |
Organism: Stigmatella aurantiaca (strain dw4/3-1)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-10-09 Classification: SIGNALING PROTEIN Ligands: EL5 |
 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Bathy Phytochrome From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-11-28 Classification: SIGNALING PROTEIN Ligands: EL5 |
 |
Crystal Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State.
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2018-11-28 Classification: FLUORESCENT PROTEIN Ligands: EL5, SO4, GOL, PEG, ETE, PG0 |

