Search Count: 26
 |
Crystal Structure Of P110Alpha-Rbd Covalently Bound To A Breaker Compound Bbo-10203 Via Cys242
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ONCOPROTEIN Ligands: A1AIR |
 |
Crystal Structure Of Wild-Type Kras (Gdp-Bound) In Complex With Mrtx849 (Adagrasib)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ONCOPROTEIN/INHIBITOR Ligands: GDP, ZN, A1B7W, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ONCOPROTEIN Ligands: GNP, MG |
 |
Crystal Structure Of Active Kras-G12C (Gmppnp-Bound) In Complex With Bbo-8520
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-12-18 Classification: ONCOPROTEIN/INHIBITOR Ligands: GNP, MG, Y8N |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2024-12-18 Classification: ONCOPROTEIN/INHIBITOR Ligands: GDP, MG, Y8N |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-03-27 Classification: PROTEIN BINDING Ligands: ZTO |
 |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2021-12-15 Classification: PROTEIN BINDING Ligands: XAY |
 |
Crystal Structure Of Pseudomonas 7A Glutaminase-Asparaginase In Complex With L-Asp At Ph 4.5
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: ASP |
 |
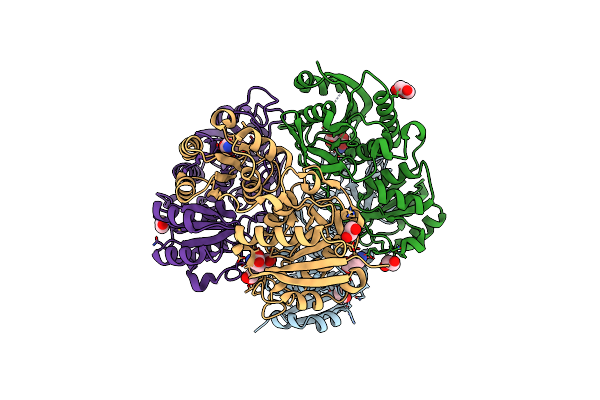
Crystal Structure Of Pseudomonas 7A Glutaminase-Asparaginase In Complex With L-Asp At Ph 5.0
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: ASP, GOL |
 |
Crystal Structure Of Pseudomonas 7A Glutaminase-Asparaginase In Complex With L-Glu At Ph 6.5
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: GLU, EDO, GOL |
 |
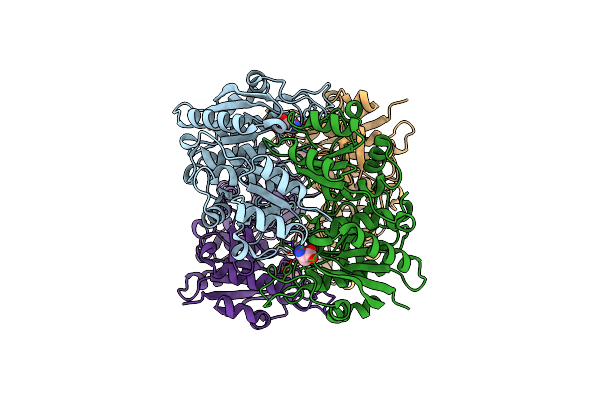
Crystal Structure Of Pseudomonas 7A Glutaminase-Asparaginase (Mutant K173M) In Complex With D-Glu At Ph 5.5
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: DGL |
 |
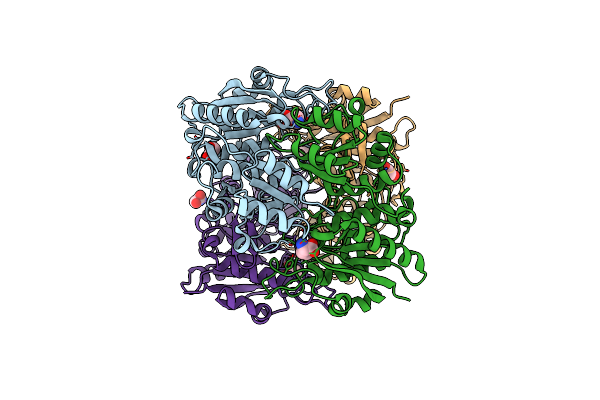
Complex Of Mutant (K173M) Of Pseudomonas 7A Glutaminase-Asparaginase With L-Asp At Ph 6. Covalent Acyl-Enzyme Intermediate
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: ASP |
 |
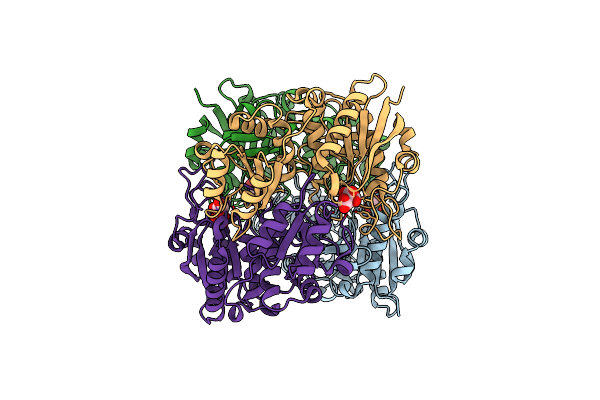
Complex Of Mutant (K173M) Of Pseudomonas 7A Glutaminase-Asparaginase With L-Glu At Ph 5. Covalent Acyl-Enzyme Intermediate
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: GLU, EDO |
 |
Complex Of Pseudomonas 7A Glutaminase-Asparaginase With Citrate Anion At Ph 5.5. Covalent Acyl-Enzyme Intermediate
Organism: Pseudomonas putida (strain atcc 47054 / dsm 6125 / ncimb 11950 / kt2440)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: CIT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-08-22 Classification: TRANSFERASE Ligands: GSH, GDS, MES, ACT, CA |
 |
Structure Of The Rpn13-Rpn2 Complex Provides Insights For Rpn13 And Uch37 As Anticancer Targets
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-04-04 Classification: PROTEIN BINDING |
 |
Two Apo Structures Of The Adenine Riboswitch Aptamer Domain Determined Using An X-Ray Free Electron Laser
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-11-23 Classification: RNA Ligands: MG |
 |
Structure Of The Adenine Riboswitch Aptamer Domain In An Intermediate-Bound State
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-11-23 Classification: RNA Ligands: ADE, MG |
 |
Ligand-Bound Structure Of Adenine Riboswitch Aptamer Domain Converted In Crystal From Its Ligand-Free State Using Ligand Mixing Serial Femtosecond Crystallography
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-11-23 Classification: RNA Ligands: ADE |

