Search Count: 205
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
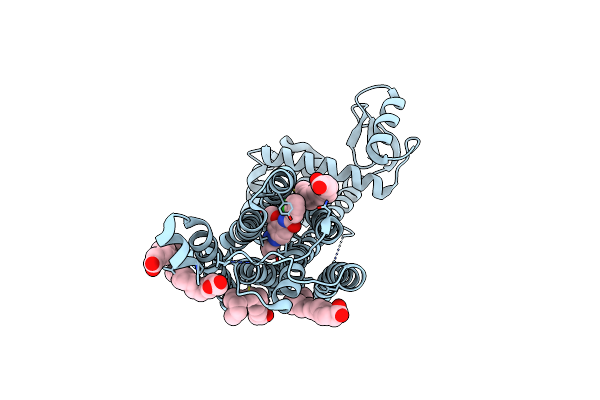 |
Dark Structure Of The Human Metabotropic Glutamate Receptor 5 Transmembrane Domain Bound To Photoswitchable Ligand Alloswitch-1
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2025-06-25 Classification: SIGNALING PROTEIN Ligands: OLA, 4YI |
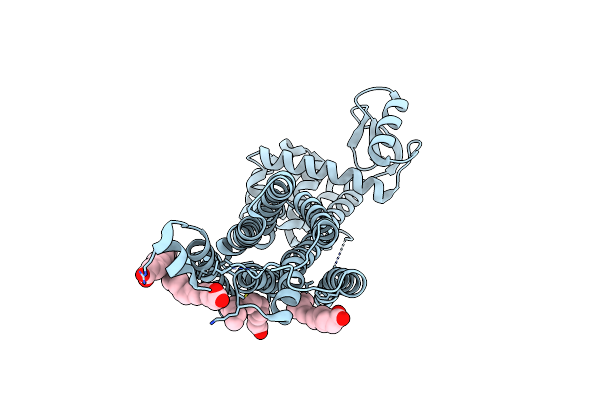 |
Apo-State Structure Of The Human Metabotropic Glutamate Receptor 5 Transmembrane Domain Freeze-Trapped After Light Activation Of Photoswitchable Ligand Alloswitch-1
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-06-25 Classification: SIGNALING PROTEIN Ligands: OLA |
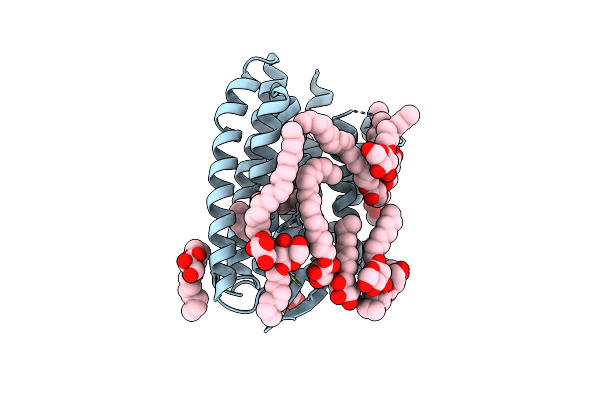 |
Grouped 150-240 Ms Dark Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL, BOG, MPG |
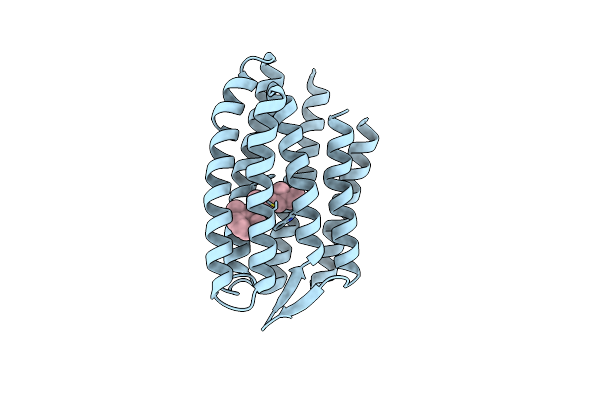 |
Continuously Illuminated Structure Of Sensory Rhodopsin Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Continuous Dark State Structure Of Sensory Rhodopsin Ii Solved By Serial Millisecond Crystallography
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-30 Classification: SIGNALING PROTEIN Ligands: RET, CL, BOG, MPG |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2025-04-02 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-03-26 Classification: PLANT PROTEIN Ligands: TLA |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 90-120 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography 60-90 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |
 |
Light Structure Of Sensory Rhodopsin-Ii Solved By Serial Millisecond Crystallography. 30-60 Milliseconds Time-Bin
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: RET, CL |

