Search Count: 72
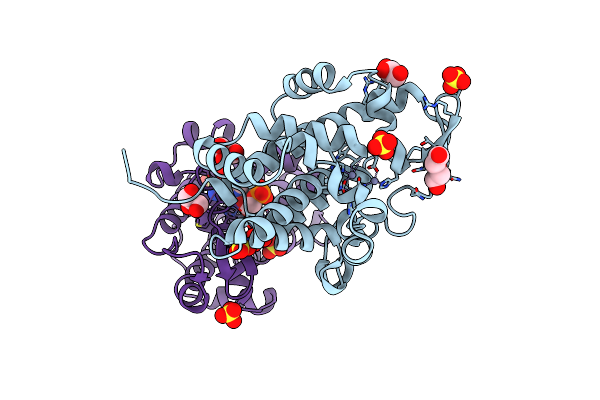 |
Smnuc1 Nuclease From Stenotrophomonas Maltophilia In Complex With Guanosine-5'-Monophosphate As A Product Of C-Di-Gmp Cleavage
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: ZN, GOL, PEG, SO4, 5GP |
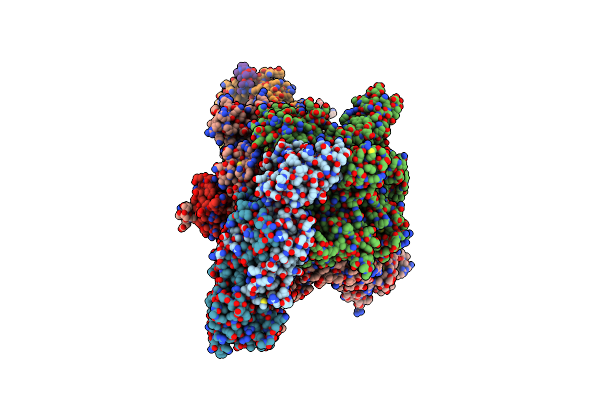 |
Mycobacterium Smegmatis Rna Polymerase In Complex With Held, Siga And Rbpa In State Ii
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: TRANSCRIPTION Ligands: ZN, MG |
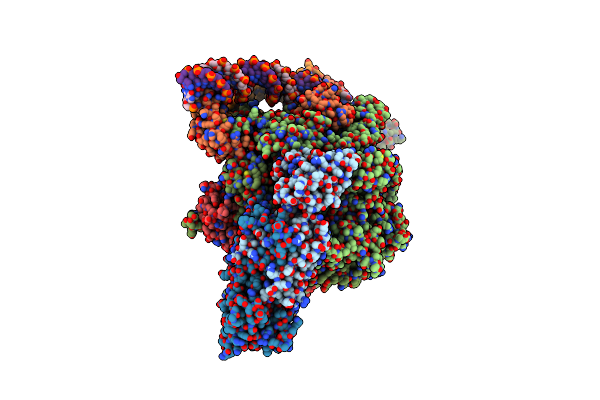 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: TRANSCRIPTION Ligands: ZN, MG |
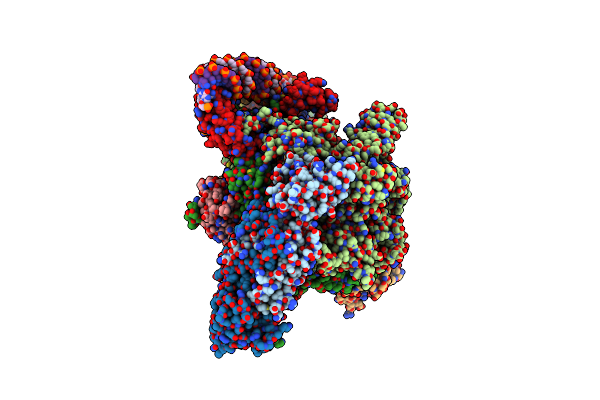 |
Mycobacterium Smegnatis Rna Polymerase Transcription Initiation Complex With Sigmaa, Rbpa, Held And An Upstream-Fork Promoter Fragment
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: TRANSCRIPTION Ligands: ZN, MG |
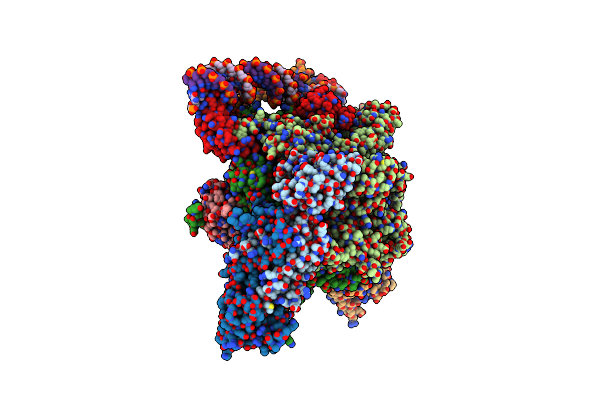 |
Mycobacterium Smegnatis Rna Polymerase Transcription Initiation Complex With Sigmaa, Rbpa, Held N-Terminal Domain And An Upstream-Fork Promoter Fragment; State Iii Conformation
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: TRANSCRIPTION Ligands: ZN, MG |
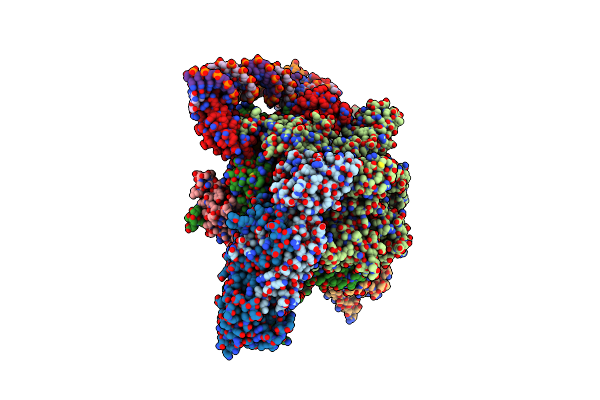 |
Mycobacterium Smegnatis Rna Polymerase Transcription Initiation Complex With Sigmaa, Rbpa, Held N-Terminal, Co And Pch Loop Domain, And An Upstream-Fork Promoter Fragment; State Iii Conformation
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: TRANSCRIPTION Ligands: ZN, MG |
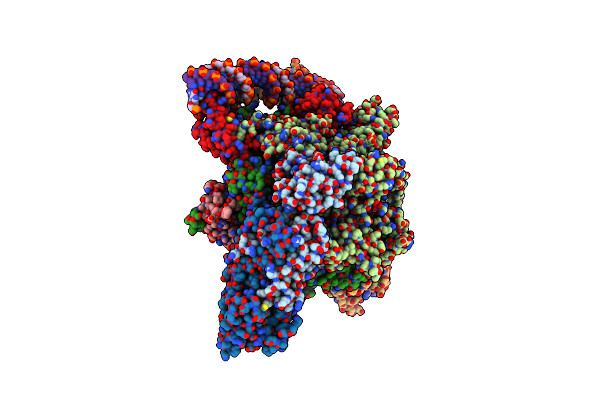 |
Mycobacterium Smegnatis Rna Polymerase Rp2-Like Transcription Initiation Complex With Sigmaa, Rbpa, Held N-Terminal Domain And Open Promoter Dna
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: TRANSCRIPTION Ligands: ZN, MG |
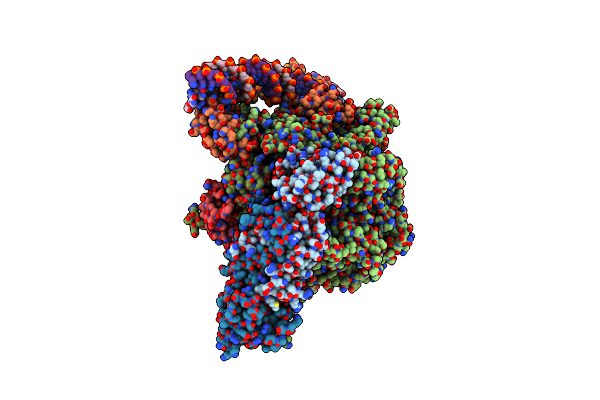 |
Mycobacterium Smegnatis Rna Polymerase Rp2-Like Transcription Initiation Complex With Sigmaa, Rbpa And Open Promoter Dna
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-09-25 Classification: HYDROLASE Ligands: ZN, GOL, SO4 |
 |
Smnuc1 Nuclease From Stenotrophomonas Maltophilia In Complex With Cytidine-5'-Monophosphate
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-09-25 Classification: HYDROLASE Ligands: C5P, ZN, SO4, NA, 1PE |
 |
Smnuc1 Nuclease From Stenotrophomonas Maltophilia In Complex With Adenosine-5'-Monophosphate
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-09-25 Classification: HYDROLASE Ligands: ZN, SO4, PO4, GOL, AMP |
 |
Smnuc1 Nuclease From Stenotrophomonas Maltophilia In Complex With Guanosine-5'-Monophosphate
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-09-25 Classification: HYDROLASE Ligands: ZN, PO4, 5GP, GOL, PEG, SO4, PGE, NA, CL |
 |
Smnuc1 Nuclease From Stenotrophomonas Maltophilia In Complex With Uridine - 5'- Monophosphate
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-09-25 Classification: HYDROLASE Ligands: ZN, GOL, U5P, PO4, SO4 |
 |
Smnuc1 Nuclease From Stenotrophomonas Maltophilia In Complex With Cytidine - 5' - Monophosphate As An Inhibitor.
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-09-25 Classification: HYDROLASE Ligands: ZN, C5P, GOL, PEG, SO4, 1PE, PO4 |
 |
Mycobacterium Smegmatis Rna Polymerase In Complex With Held, Siga And Rbpa In State I
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: FAD, EDO, SO4, NO3, PEG, CL |
 |
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: ZN, NAG, PO4, URI, NA, BTB |
 |
S1 Nuclease From Aspergillus Oryzae In Complex With Cytidine-5'-Monophosphate
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: ZN, NAG, PO4, C5P, BTB, CA |
 |
Organism: Paenibacillus thiaminolyticus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: TBR, ZN, GOL, SO4 |
 |
Organism: Chaetomium thermophilum var. thermophilum dsm 1495
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2021-05-26 Classification: OXIDOREDUCTASE Ligands: FDA, NAG, FMT, MG |

