Search Count: 11
 |
Klentaq Dna Polymerase Processing A Modified Primer - Bearing The Modification At The 3'-Terminus Of The Primer.
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-09-26 Classification: DNA BINDING PROTEIN Ligands: XG4, EDO, MN, MG |
 |
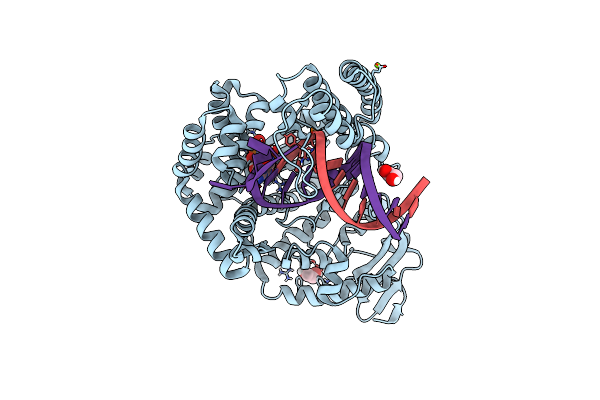
Klentaq Dna Polymerase Processing A Modified Primer - Bearing The Modification Upstream At The Second Primer Nucleotide.
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-09-26 Classification: DNA BINDING PROTEIN Ligands: MN, XG4, MG, GOL |
 |
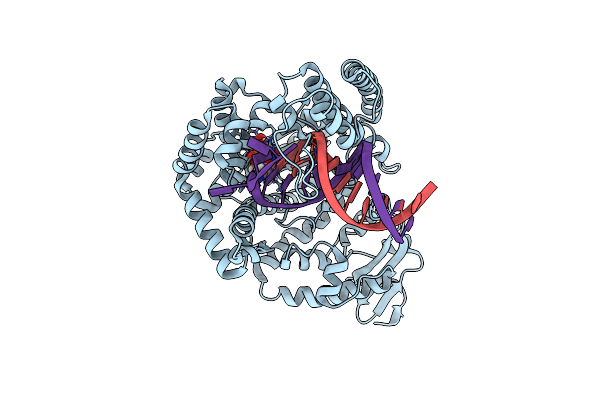
Klentaq Dna Polymerase Processing A Modified Primer - Bearing The Modification Upstream At The Third Primer Nucleotide.
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2018-09-26 Classification: DNA BINDING PROTEIN Ligands: MN, EDO, XG4, MG |
 |
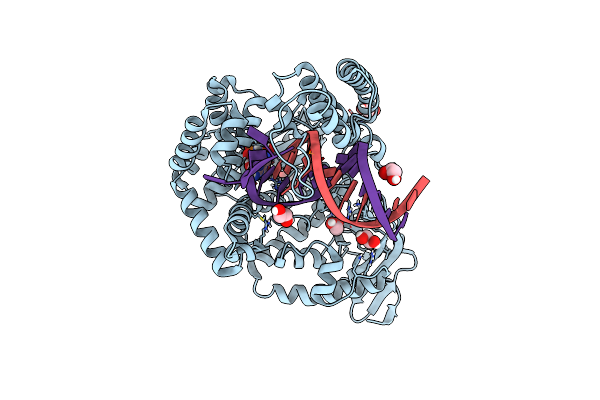
Klentaq Dna Polymerase Processing A Modified Primer - Bearing The Modification Upstream At The Fourth Primer Nucleotide.
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-09-26 Classification: DNA BINDING PROTEIN Ligands: MN, XG4 |
 |
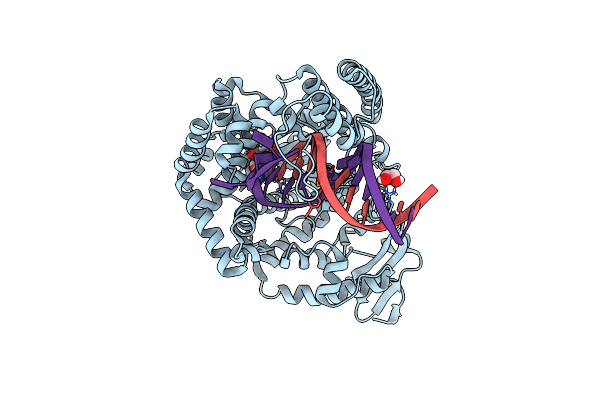
Klentaq Dna Polymerase Processing A Modified Primer - Bearing The Modification Upstream At The Fifth Primer Nucleotide.
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-09-26 Classification: DNA BINDING PROTEIN Ligands: MN, EDO, XG4 |
 |
Klentaq Dna Polymerase Processing A Modified Primer - Bearing The Modification Upstream At The Sixth Primer Nucleotide.
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-09-26 Classification: DNA BINDING PROTEIN Ligands: MN, EDO, XG4 |
 |
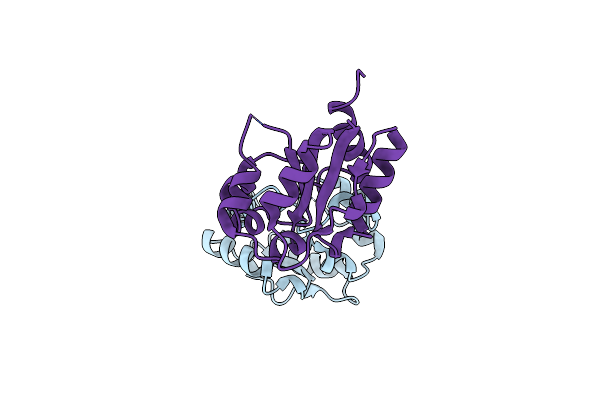
Klentaq Dna Polymerase In A Closed, Ternary Complex With Dgpnhpp Bound In The Active Site
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-09-26 Classification: DNA BINDING PROTEIN Ligands: XG4, MN, EDO |
 |
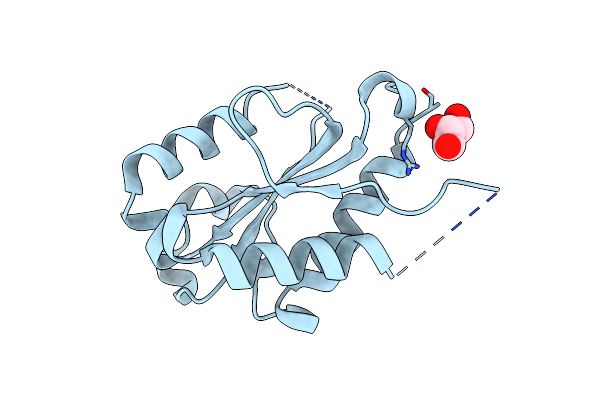
The Crystal Structures Of The Brucella Protein Tcpb And The Tlr Adaptor Protein Tirap Show Structural Differences In Microbial Tir Mimicry.
Organism: Brucella melitensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-12-04 Classification: IMMUNE SYSTEM |
 |
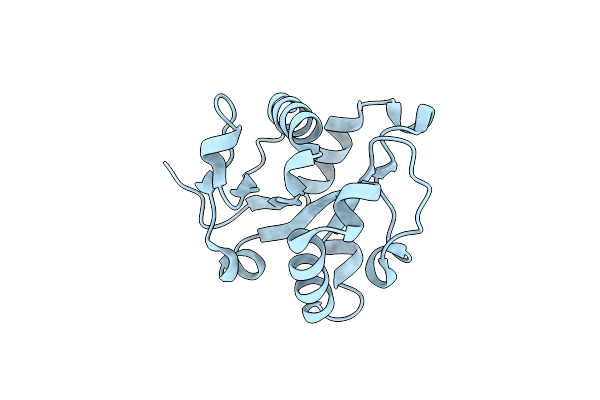
The Crystal Structures Of The Brucella Protein Tcpb And The Tlr Adaptor Protein Tirap Show Structural Differences In Microbial Tir Mimicry
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2013-12-04 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
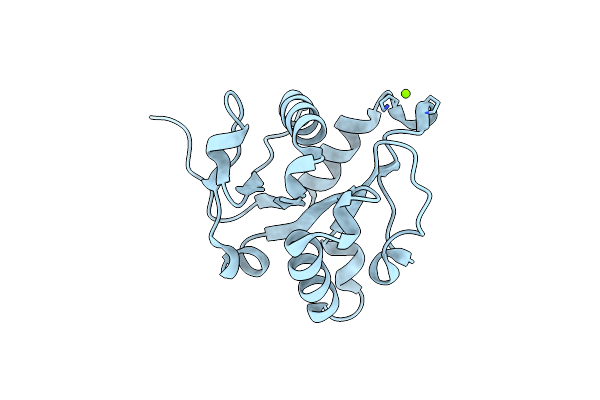
Crystal Structure Of The Tir-Domain Of Human Myeloid Differentiation Primary Response Protein (Myd88)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-04-10 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of The Tir Domain Of Human Myeloid Differentiation Primary Response Protein 88.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2013-04-10 Classification: PROTEIN BINDING Ligands: MG |

