Search Count: 56
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2020-01-08 Classification: RNA Ligands: MG, K |
 |
X-Ray Crystal Structure Of The Tetrahydrofolate Riboswitch Aptamer Bound To 5-Deazatetrahydropterin
Organism: Streptococcus mutans ua159
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2019-12-18 Classification: RNA Ligands: T0A, MG |
 |
Crystal Structure Of Bh32 Alkylated With The Mechanistic Inhibitor 2-Bromoacetophenone
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: AC0 |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: CA |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: PGE, AC0, SO4, EDO, MG |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-06-05 Classification: HYDROLASE |
 |
Crystal Structure Of Oe1.3 Alkylated With The Mechanistic Inhibitor 2-Bromoacetophenone
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-06-05 Classification: HYDROLASE Ligands: AC0, ACT, PEG |
 |
Crystal Structure Of Coclaurine N-Methyltransferase (Cnmt) Bound To N-Methylheliamine And Sah
Organism: Coptis japonica
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: SAH |
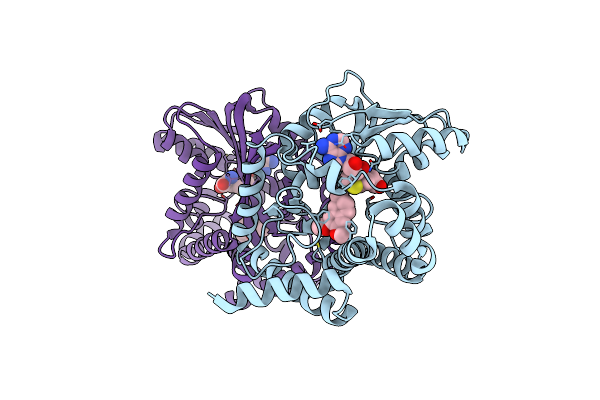 |
Crystal Structure Of Coclaurine N-Methyltransferase (Cnmt) Bound To N-Methylheliamine And Sah
Organism: Coptis japonica
Method: X-RAY DIFFRACTION Release Date: 2018-06-06 Classification: TRANSFERASE Ligands: SAH, F2W |
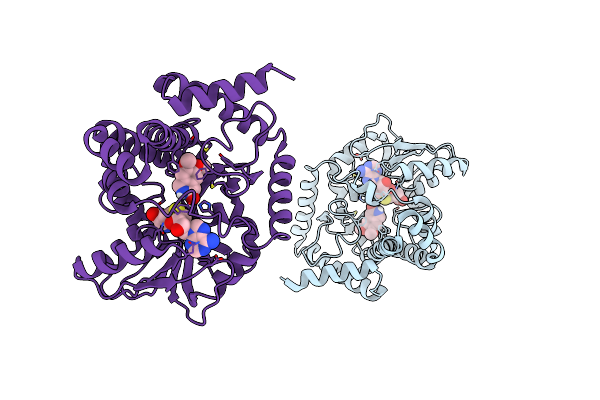 |
Crystal Structure Of Coclaurine N-Methyltransferase (Cnmt) Bound To N-Methylheliamine And Sah
Organism: Coptis japonica
Method: X-RAY DIFFRACTION Release Date: 2018-06-06 Classification: TRANSFERASE Ligands: SAH, F2Z |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2018-05-23 Classification: ISOMERASE |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-05-23 Classification: ISOMERASE |
 |
Crystal Structure Of Ketosteroid Isomerase Quadruple Variant V88I/L99V/D103S/V101A
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2018-05-23 Classification: ISOMERASE |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2018-05-23 Classification: ISOMERASE |
 |
Structure Of The A Domain Of Carboxylic Acid Reductase (Car) From Nocardia Iowensis In Complex With Amp And Iodide
Organism: Nocardia iowensis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2018-01-17 Classification: OXIDOREDUCTASE Ligands: AMP, IOD |
 |
Crystal Structure Of Phenylalanine Ammonia-Lyase From Anabaena Variabilis (Y78F-C503S-C565S) Bound To Cinnamate
Organism: Anabaena
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2017-09-20 Classification: LYASE Ligands: HCI |
 |
Crystal Structure Of The Engineered D-Amino Acid Dehydrogenase (Daadh) Bound To Nadp+
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2017-08-16 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2017-08-16 Classification: OXIDOREDUCTASE |
 |
Structure Of The A Domain Of Carboxylic Acid Reductase (Car) From Nocardia Iowensis In Complex With Amp
Organism: Nocardia iowensis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-07-05 Classification: OXIDOREDUCTASE Ligands: AMP |
 |
Structure Of The A Domain Of Carboxylic Acid Reductase (Car) From Nocardia Iowensis In Complex With Amp And Benzoic Acid
Organism: Nocardia iowensis
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2017-07-05 Classification: OXIDOREDUCTASE Ligands: AMP, BEZ |

