Search Count: 53
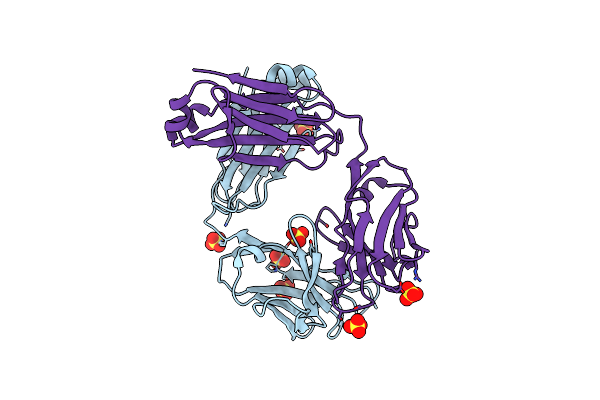 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-07-03 Classification: IMMUNE SYSTEM Ligands: SO4 |
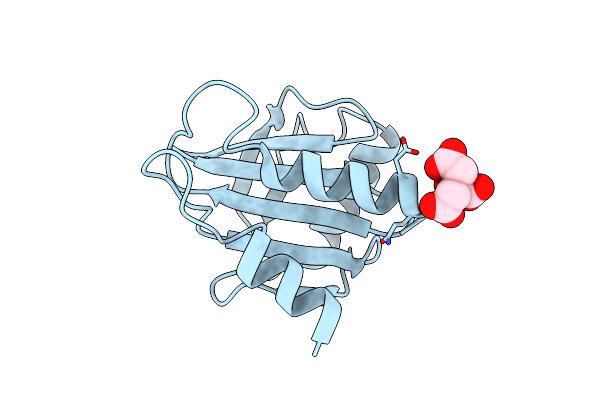 |
Crystal Structure Of Timothy Grass Allergen Phl P 12.0101 Reveals An Unusual Profilin Dimer
Organism: Phleum pratense
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-03-31 Classification: ALLERGEN Ligands: CIT |
 |
Organism: Geobacter sulfurreducens (strain atcc 51573 / dsm 12127 / pca)
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2020-02-19 Classification: ELECTRON TRANSPORT Ligands: SO4, HEM |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-11-20 Classification: IMMUNE SYSTEM Ligands: CL, FMT, ACT |
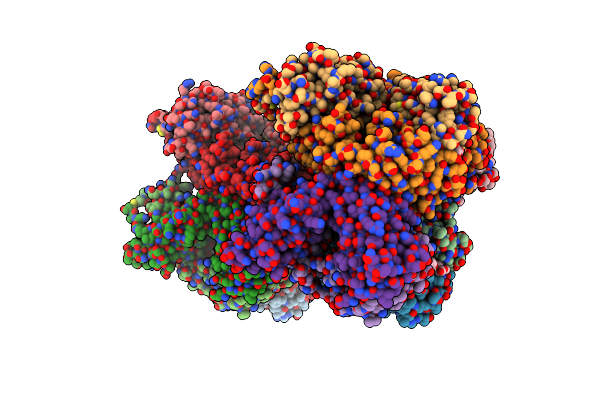 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2019-11-20 Classification: IMMUNE SYSTEM Ligands: BCG, PEG, GOL, ACT, FMT |
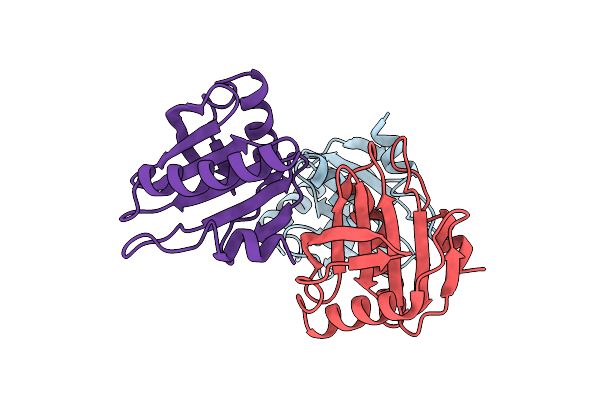 |
Organism: Cucumis melo
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-08-21 Classification: ALLERGEN |
 |
Organism: Artemisia vulgaris
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-10-03 Classification: ALLERGEN |
 |
Crystal Structure Of Rimk Domain Protein Atp-Grasp From Sphaerobacter Thermophilus Dsm 20745
Organism: Sphaerobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2016-03-16 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
 |
Crystal Structure Of O-Methyltransferase Family 2 Protein Plim_1147 From Planctomyces Limnophilus Dsm 3776 Complex With Apigenin
Organism: Planctopirus limnophila (strain atcc 43296 / dsm 3776 / ifam 1008 / 290)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2016-03-02 Classification: TRANSFERASE Ligands: AGI, EDO, FMT |
 |
Crystal Structure Of Unknown Function Protein Dfer_1899 Fromdyadobacter Fermentans Dsm 18053
Organism: Dyadobacter fermentans (strain atcc 700827 / dsm 18053 / ns114)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-02-17 Classification: Structural Genomics, Unknown Function Ligands: PG4, ACT, GOL, PGE |
 |
Crystal Structure Of Known Function Protein From Kribbella Flavida Dsm 17836
Organism: Kribbella flavida (strain dsm 17836 / jcm 10339 / nbrc 14399)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-12-02 Classification: UNKNOWN FUNCTION Ligands: GOL |
 |
Crystal Structure Of Abc Transporter System Solute-Binding Protein From Rhodopirellula Baltica Sh 1
Organism: Rhodopirellula baltica (strain dsm 10527 / ncimb 13988 / sh1)
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2015-11-25 Classification: solute-binding protein Ligands: CA, GOL |
 |
Organism: Geobacter sulfurreducens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-04-01 Classification: ELECTRON TRANSPORT Ligands: HEM, ZN, CAC, ACT |
 |
Organism: Anaeromyxobacter dehalogenans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-07-17 Classification: SIGNALING PROTEIN Ligands: ZN, ACT |
 |
Organism: Anaeromyxobacter dehalogenans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-06-12 Classification: SIGNALING PROTEIN Ligands: ZN, CL, NA, ACT |
 |
Mitigation Of X-Ray Damage In Macromolecular Crystallography By Submicrometer Line Focusing; Total Dose 2.17 X 10E+12 X-Ray Photons
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, NA, EDO |
 |
Mitigation Of X-Ray Damage In Macromolecular Crystallography By Submicrometer Line Focusing; Total Dose 1.32 X 10E+12 X-Ray Photons
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, NA, EDO |
 |
Mitigation Of X-Ray Damage In Macromolecular Crystallography By Submicrometer Line Focusing; Total Dose 6.70 X 10E+11 X-Ray Photons
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: CL, NA, EDO |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-08-03 Classification: Structural Genomics, Unknown Function Ligands: GOL |
 |
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-06-01 Classification: HYDROLASE Ligands: NAG |

