Search Count: 15
 |
Structural Basis Of L-Phosphoserine Binding To Bacillus Alcalophilus Phosphoserine Aminotransferase
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-05-01 Classification: TRANSFERASE Ligands: SEP, PLP, CL, NA |
 |
Structural Basis Of L-Phosphoserine Binding To Bacillus Alcalophilus Phosphoserine Aminotransferase
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-05-01 Classification: TRANSFERASE Ligands: PLP, CL |
 |
Conserved Hydrophobic Clusters On The Surface Of The Caf1A Usher C-Terminal Domain Are Important For F1 Antigen Assembly
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-09-22 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Phosphoserine Aminotransferase From Bacillus Circulans Var. Alkalophilus At Ph 8.5
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2006-03-22 Classification: TRANSFERASE Ligands: PLP |
 |
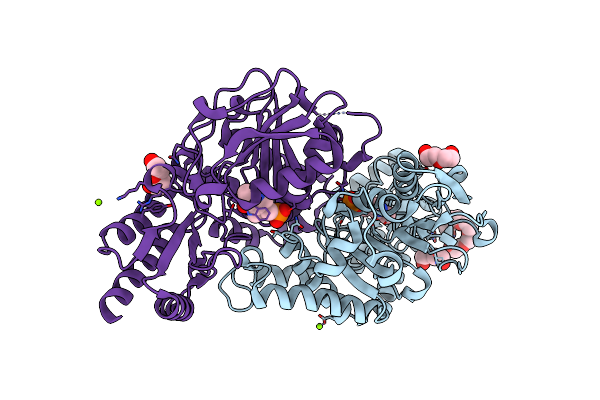
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure A)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
 |
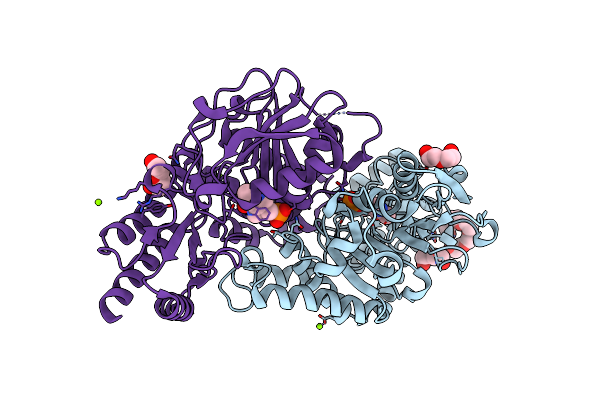
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure B)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
 |
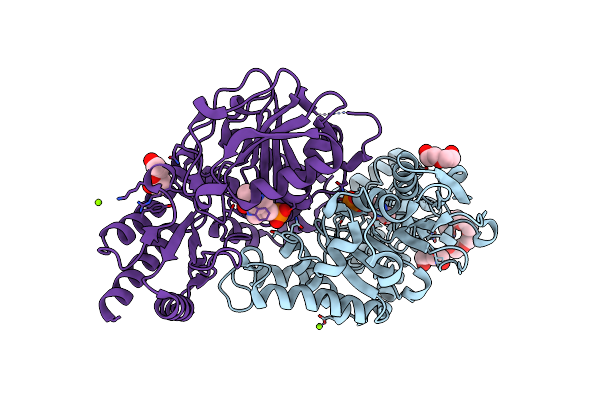
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure C)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
 |
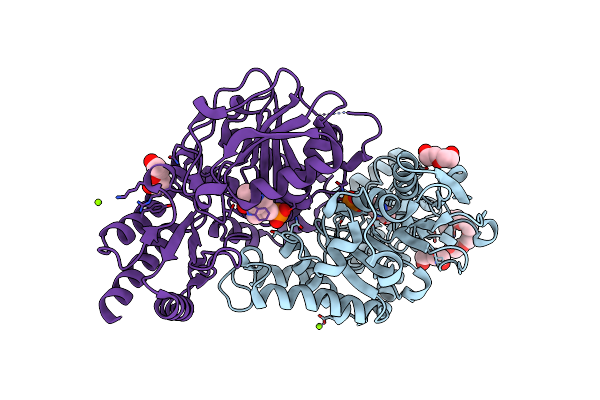
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure D)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
 |
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure E)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
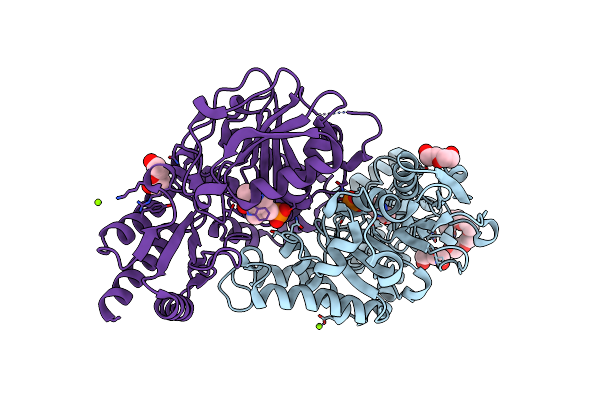 |
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure F)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
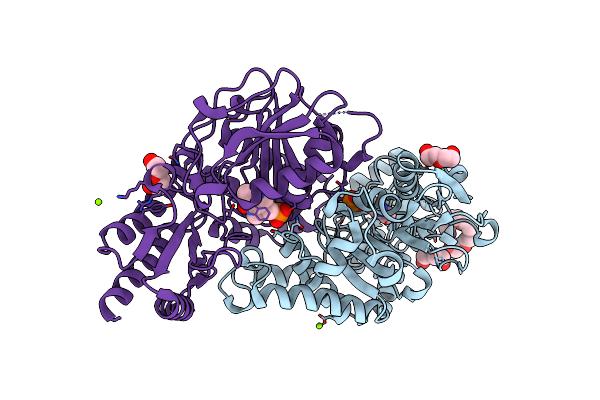 |
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure G)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
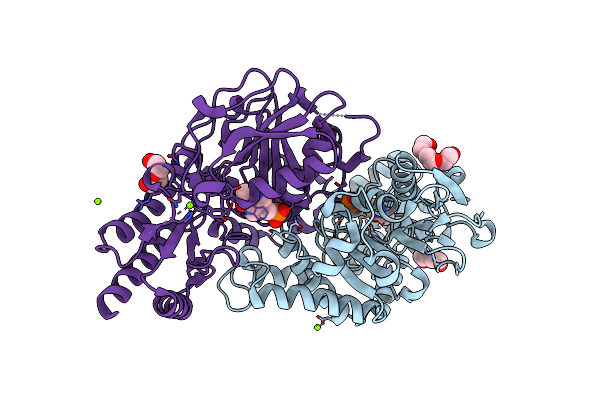 |
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure H)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, PGE, PEG |
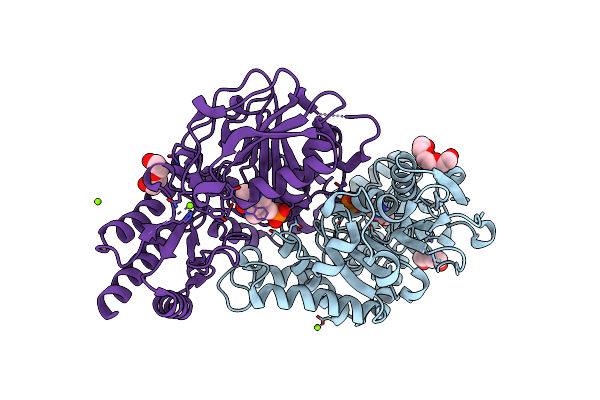 |
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure I)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, PGE, PEG |
 |
Crystal Structure Of Phosphoserine Aminotransferase From Bacillus Circulans Var. Alkalophilus
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2004-12-22 Classification: TRANSFERASE Ligands: PLP, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2002-10-23 Classification: IMMUNE SYSTEM |

