Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
1.65 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) From Staphylococcus Aureus With Bme-Modified Cys289 And Peg Molecule In Active Site
Organism: Staphylococcus aureus subsp. aureus col
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-07-16 Classification: OXIDOREDUCTASE |
 |
2.6 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) G234S Mutant From Staphylococcus Aureus (Idp00699) In Complex With Nad+ And Bme-Free Cys289
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-07-02 Classification: OXIDOREDUCTASE Ligands: NAD, ACT |
 |
1.85 Angstrom Resolution Crystal Structure Of Apo Betaine Aldehyde Dehydrogenase (Betb) G234S Mutant From Staphylococcus Aureus (Idp00699) With Bme-Free Sulfinic Acid Form Of Cys289
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-06-18 Classification: OXIDOREDUCTASE |
 |
1.90 Angstrom Resolution Crystal Structure Of Apo Betaine Aldehyde Dehydrogenase (Betb) G234S Mutant From Staphylococcus Aureus (Idp00699) With Bme-Modified Cys289
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-05-07 Classification: OXIDOREDUCTASE |
 |
2.30 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) From Staphylococcus Aureus With Bme-Free Cys289
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-12-11 Classification: OXIDOREDUCTASE |
 |
1.90 Angstrom Resolution Crystal Structure Of Betaine Aldehyde Dehydrogenase (Betb) From Staphylococcus Aureus In Complex With Nad+ And Bme-Free Cys289
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-11-06 Classification: OXIDOREDUCTASE Ligands: NAD, K |
 |
1.95 Angstrom Crystal Structure Of Of Type I 3-Dehydroquinate Dehydratase (Arod) From Clostridium Difficile With Covalent Modified Comenic Acid.
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-09-26 Classification: LYASE Ligands: PGE, PEG, SHL, ACT |
 |
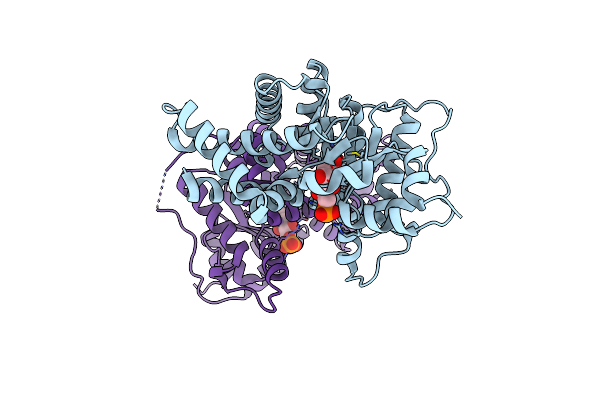
1.8 Angstrom Resolution Crystal Structure Of Transaldolase From Francisella Tularensis (Phosphate-Free)
Organism: Francisella tularensis subsp. tularensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-03-14 Classification: TRANSFERASE Ligands: MG, ACT |
 |
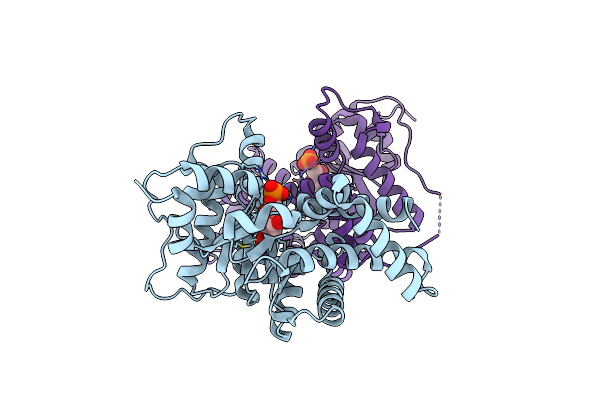
1.65 Angstrom Resolution Crystal Structure Of Transaldolase B (Tala) From Francisella Tularensis In Covalent Complex With Sedoheptulose-7-Phosphate
Organism: Francisella tularensis subsp. tularensis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2011-09-14 Classification: TRANSFERASE Ligands: I22, CL |
 |
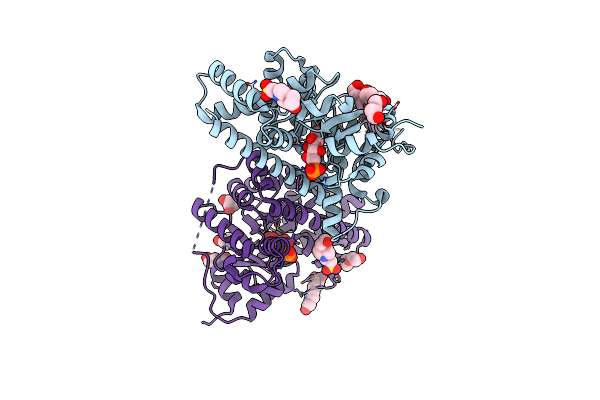
2.0 Angstrom Resolution Crystal Structure Of Transaldolase B (Tala) From Francisella Tularensis In Covalent Complex With Fructose 6-Phosphate
Organism: Francisella tularensis subsp. tularensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-09-07 Classification: TRANSFERASE Ligands: F6R |
 |
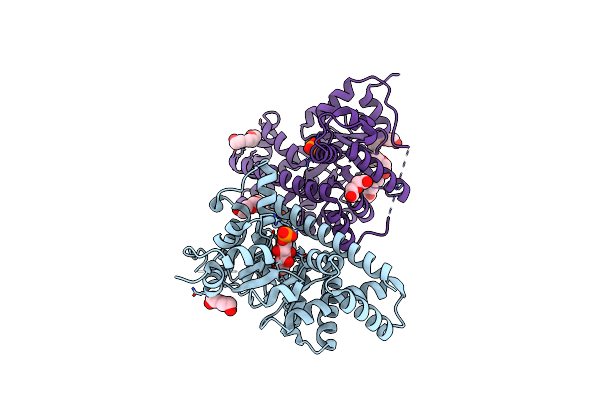
1.5 Angstrom Resolution Crystal Structure Of K135M Mutant Of Transaldolase B (Tala) From Francisella Tularensis In Complex With Sedoheptulose 7-Phosphate.
Organism: Francisella tularensis subsp. tularensis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-09-07 Classification: TRANSFERASE Ligands: I22, EPE, PEG, GOL |
 |
1.8 Angstrom Resolution Crystal Structure Of K135M Mutant Of Transaldolase B (Tala) From Francisella Tularensis In Complex With Fructose 6-Phosphate
Organism: Francisella tularensis subsp. tularensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-08-24 Classification: TRANSFERASE Ligands: F6R, PEG, PGE, PO4 |
 |
Crystal Structure Of The 3-Dehydroquinate Dehydratase (Arod) From Salmonella Enterica Typhimurium Lt2 With Malonate And Boric Acid At The Active Site
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2011-06-01 Classification: LYASE Ligands: MLA, DMS, NI, IMD, BO3 |
 |
Crystal Structure Of K170M Mutant Of Type I 3-Dehydroquinate Dehydratase (Arod) From Salmonella Typhimurium Lt2 In Non-Covalent Complex With Dehydroquinate.
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-07-28 Classification: LYASE Ligands: DQA |
 |
Crystal Structure Of Type I 3-Dehydroquinate Dehydratase (Arod) From Salmonella Typhimurium Lt2 In Covalent Complex With Dehydroquinate
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2010-04-07 Classification: LYASE Ligands: DQA, GOL |
 |
Crystal Structure Of Type I 3-Dehydroquinate Dehydratase (Arod) From Clostridium Difficile With Covalent Reaction Intermediate
Organism: Clostridium difficile
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-09-22 Classification: LYASE Ligands: DHS |
 |
Organism: Bacillus anthracis str.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-06-19 Classification: HYDROLASE |
 |
Organism: Bacillus anthracis str.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-06-19 Classification: HYDROLASE Ligands: ACY |