Search Count: 13
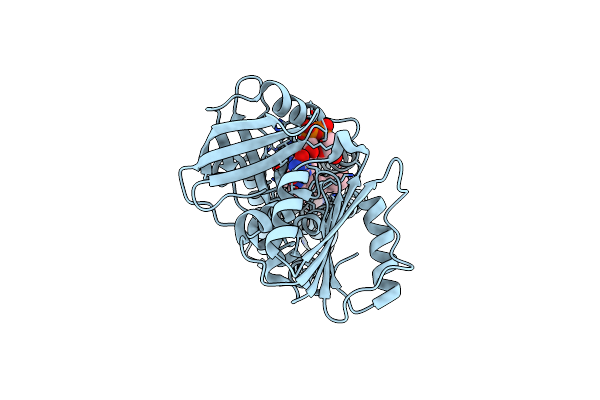 |
Organism: Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: MEMBRANE PROTEIN Ligands: FAD, HEM |
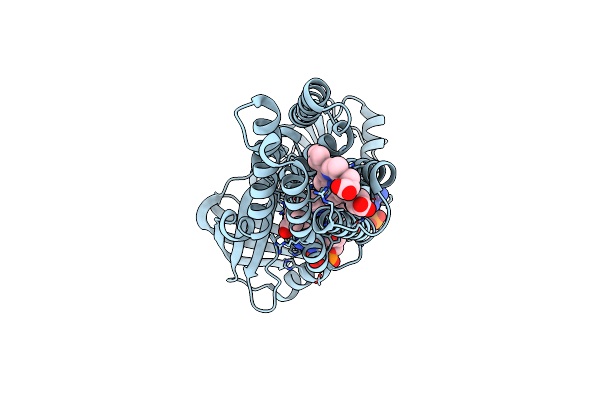 |
Cryo-Em Structure Of Streptococcus Pneumoniae Nadph Oxidase In Complex With Nadph
Organism: Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: MEMBRANE PROTEIN Ligands: NDP, FAD, HEM |
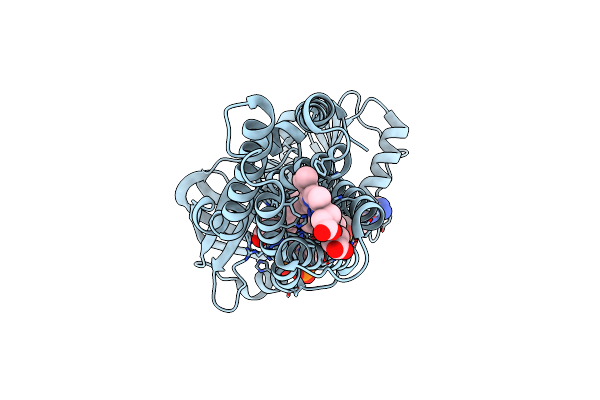 |
Cryo-Em Structure Of Stably Reduced Streptococcus Pneumoniae Nadph Oxidase In Complex With Nadh
Organism: Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: MEMBRANE PROTEIN Ligands: FAD, HEM, NAI |
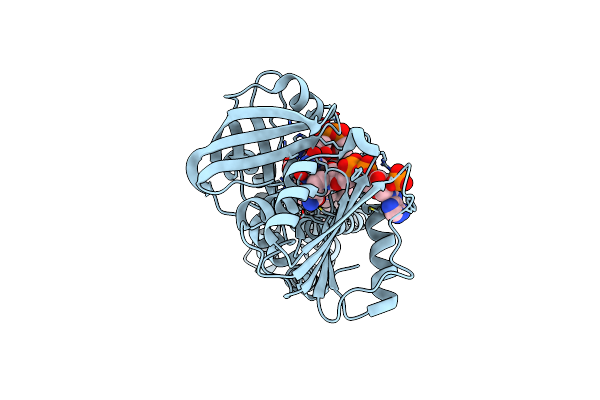 |
Cryo-Em Structure Of Streptococcus Pneumoniae Nadph Oxidase F397A Mutant In Complex With Nadph
Organism: Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: MEMBRANE PROTEIN Ligands: NDP, FAD, HEM |
 |
Cryo-Em Map Of The Wt Kdpfabc Complex In The E1-P Tight Conformation, Stabilised With The Inhibitor Orthovanadate
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: K, CDL, VO4 |
 |
Cryo-Em Map Of The Wt Kdpfabc Complex In The E1-P Tight Conformation, Under Turnover Conditions
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: K, CDL |
 |
Cryo-Em Map Of The Wt Kdpfabc Complex In The E1_Atpearly Conformation, Under Turnover Conditions
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: K, CDL, ATP |
 |
Cryo-Em Structure Of The Kdpfabc Complex In A Nucleotide-Free E1 Conformation Loaded With K+
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: K, CDL |
 |
Cryo-Em Structure Of The Kdpfabc Complex In A Nucleotide-Free E1 Conformation Loaded With K+
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: K, CDL |
 |
Cryo-Em Structure Of The Kdpfabc Complex In A Nucleotide-Free E1 Conformation Loaded With K+
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: K, CDL |
 |
Cryo-Em Map Of The Wt Kdpfabc Complex In The E1-P_Adp Conformation, Under Turnover Conditions
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: K, CDL, ADP |
 |
Cryo-Em Map Of The Unphosphorylated Kdpfabc Complex In The E2-P Conformation, Under Turnover Conditions
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Cryo-Em Map Of The Unphosphorylated Kdpfabc Complex In The E1-P_Adp Conformation, Under Turnover Conditions
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: MEMBRANE PROTEIN Ligands: K, ADP, MG |

