Search Count: 17
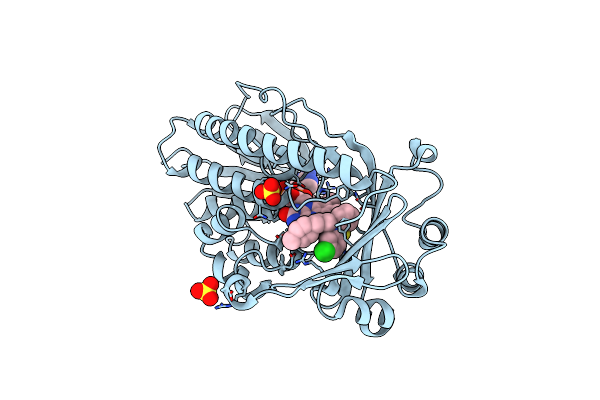 |
Crystal Structure Of Flavin-Dependent Monooxygenase Tet(X4) In Complex With Prochlorperazine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: FDA, P77, SO4 |
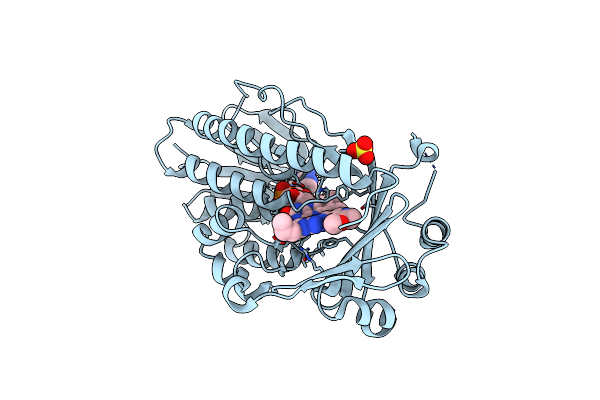 |
Crystal Structure Of Flavin-Dependent Monooxygenase Tet(X4) In Complex With Tegaserod
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: FDA, A1IVL, SO4 |
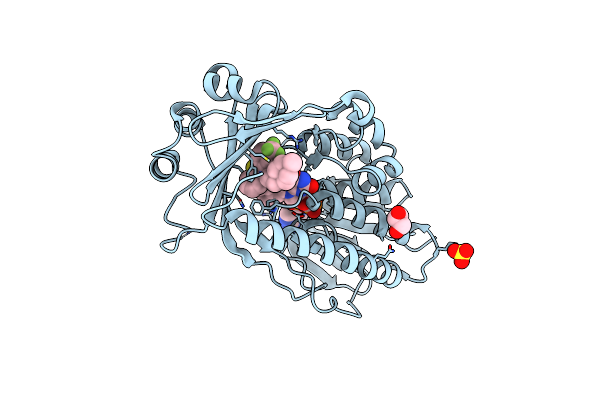 |
Crystal Structure Of Flavin-Dependent Monooxygenase Tet(X4) In Complex With Trifluoperazine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: FDA, GOL, TFP, SO4 |
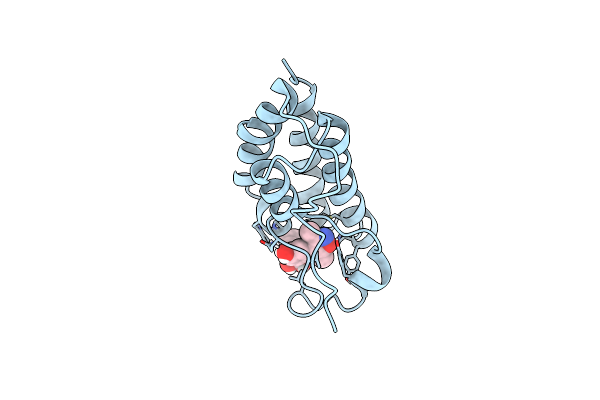 |
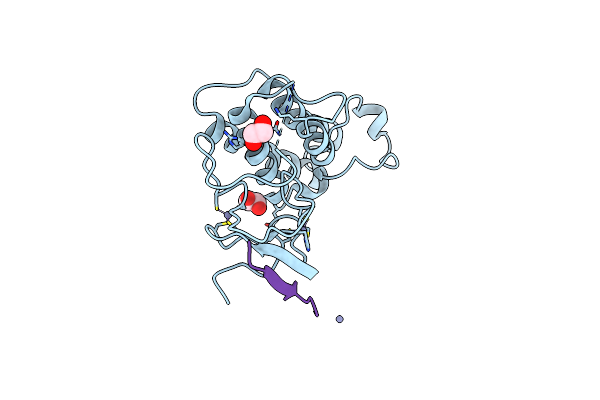
Crystal Structure Of The First Bromodomain Of Human Brd4 L94C Variant In Complex With Racemic 3,5-Dimethylisoxazol Ligand
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-08-02 Classification: GENE REGULATION Ligands: E5Q, V0R |
 |
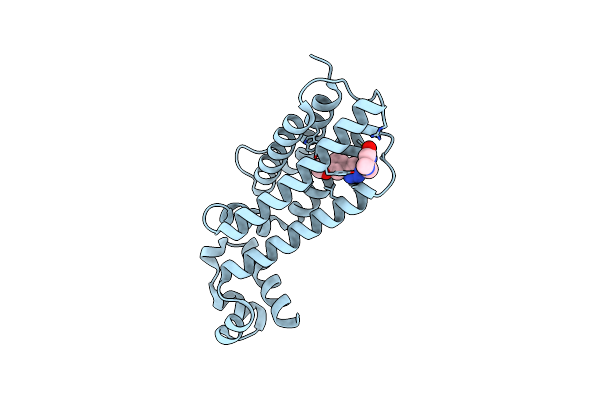
Crystal Structure Of Trim33 Phd-Bromodomain Isoform B In Complex With H3K10Ac Histone Peptide.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-06-29 Classification: TRANSCRIPTION Ligands: EDO, ZN |
 |
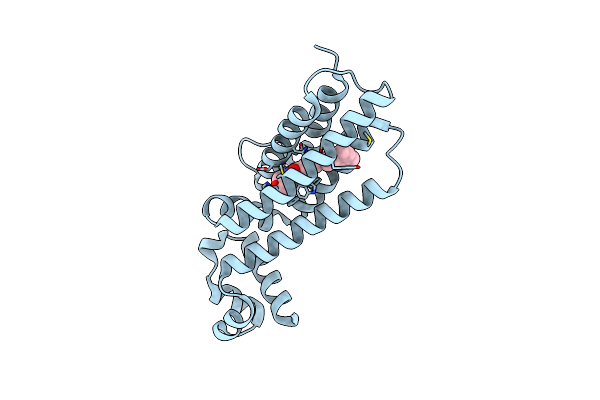
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-19 Classification: DNA BINDING PROTEIN Ligands: JPH |
 |
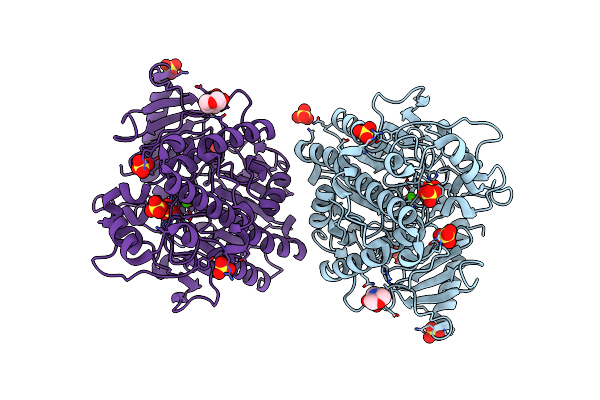
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-19 Classification: DNA BINDING PROTEIN Ligands: JPK |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-03-16 Classification: HYDROLASE Ligands: CA, SO4, MES, CL |
 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-09-16 Classification: HYDROLASE |
 |
G4 Mutant Of Pas, Arylsulfatase From Pseudomonas Aeruginosa, In Complex With Phenylphosphonic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-05-13 Classification: HYDROLASE Ligands: CA, SO4, SV7 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2015-04-29 Classification: HYDROLASE Ligands: PEG, SO4, CA |
 |
G6 Mutant Of Pas, Arylsulfatase From Pseudomonas Aeruginosa, In Complex With Phenylphosphonic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2015-04-29 Classification: HYDROLASE Ligands: CA, SV7, PEG, NH4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2015-04-22 Classification: HYDROLASE Ligands: CA, SO4, PEG |
 |
G4 Mutant Of Pas, Arylsulfatase From Pseudomonas Aeruginosa, In Complex With 3-Br-Phenolphenylphosphonate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2015-04-22 Classification: HYDROLASE Ligands: CA, 62Y |
 |
Novel Aromatic Inhibitors Of Influenza Virus Neuraminidase Make Selective Interactions With Conserved Residues And Water Molecules In The Active Site
Organism: Influenza b virus (b/lee/40)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1999-02-27 Classification: HYDROLASE Ligands: NAG, CA, RAI |
 |
Novel Aromatic Inhibitors Of Influenza Virus Neuraminidase Make Selective Interactions With Conserved Residues And Water Molecules In Teh Active Site
Organism: Influenza b virus (b/lee/40)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 1999-02-27 Classification: HYDROLASE Ligands: NAG, CA, RA2 |
 |
Novel Aromatic Inhibitors Of Influenza Virus Neuraminidase Make Selective Interactions With Conserved Residues And Water Molecules In The Active Site
Organism: Influenza b virus (b/lee/40)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1999-02-26 Classification: HYDROLASE Ligands: NAG, CA, FDI |

