Search Count: 54
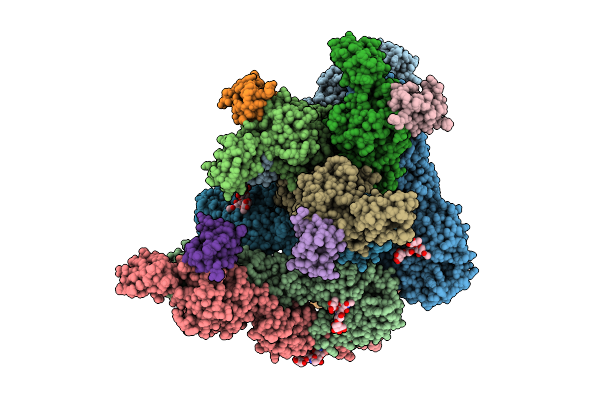 |
Organism: Chikungunya virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: VIRUS |
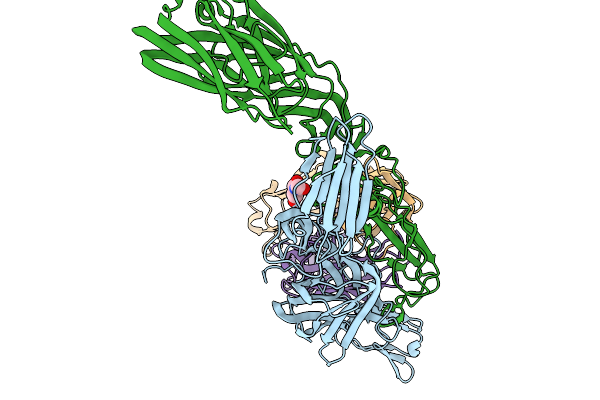 |
Organism: Chikungunya virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: ANTIFUNGAL PROTEIN Ligands: NAG |
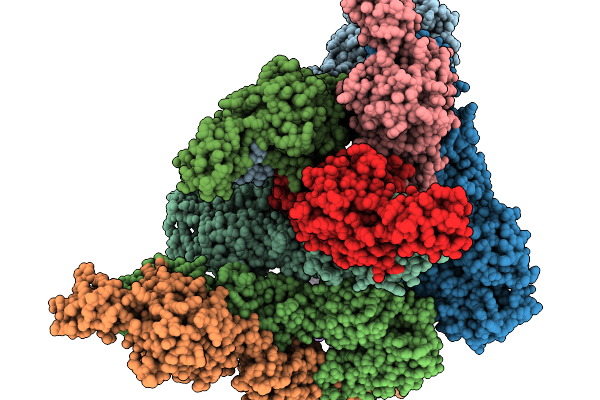 |
Organism: Chikungunya virus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRUS LIKE PARTICLE |
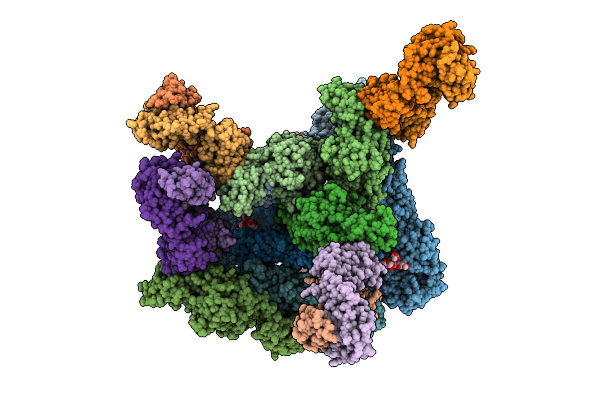 |
Organism: Homo sapiens, Chikungunya virus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRUS LIKE PARTICLE Ligands: CLR |
 |
Organism: Chikungunya virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ANTIVIRAL PROTEIN |
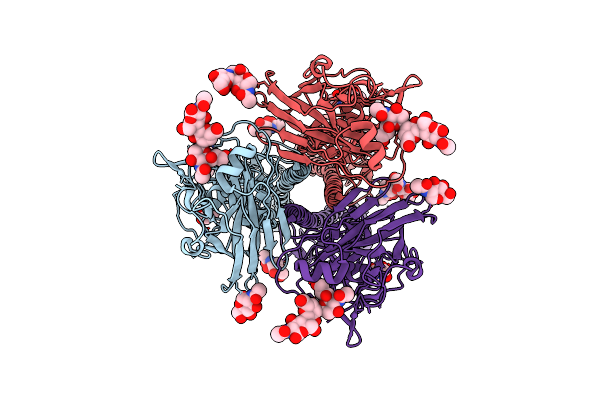 |
Cryo-Em Structure Of A Human-Infecting Bovine Influenza H5N1 Hemagglutinin Complexed With Avian Receptor Analog Lsta
Organism: Influenza a virus (a/indonesia/5/2005(h5n1))
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: NAG, SIA |
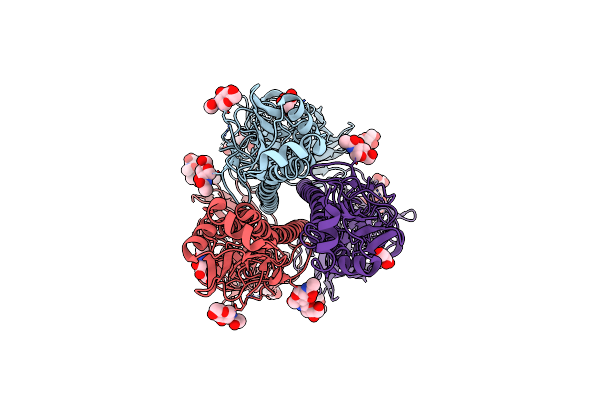 |
Cryo-Em Structure Of A Human-Infecting Bovine Influenza H5N1 Hemagglutinin Complexed With Human Receptor Analog Lstc
Organism: Influenza a virus (a/indonesia/5/2005(h5n1))
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: NAG |
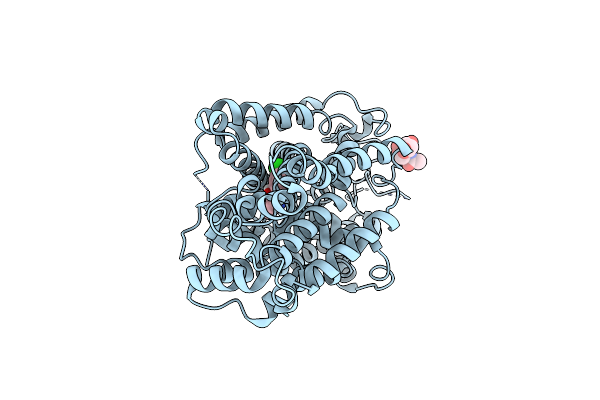 |
Structure Of Radafaxine-Bound State Of The Human Norepinephrine Transporter
Organism: Homo sapiens, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: TRANSPORT PROTEIN Ligands: NAG, YNT |
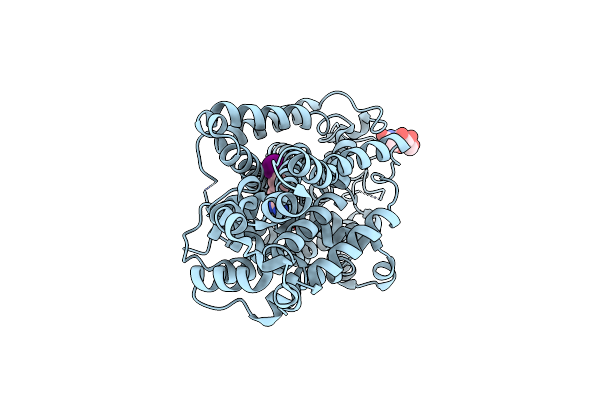 |
Structural Mechanism Of Substrate Binding And Inhibition Of The Human Norepinephrine Transporter
Organism: Homo sapiens, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: TRANSPORT PROTEIN Ligands: YMN, NAG |
 |
Organism: Homo sapiens, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: TRANSPORT PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Thehydroxycarboxylic Acid Receptor 2-Gi Protein Complex Bound Mk-6892
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: FI7 |
 |
Cryo-Em Structure Of Thehydroxycarboxylic Acid Receptor 2-Gi Protein Complex Bound Niacin
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: MEMBRANE PROTEIN Ligands: NIO |
 |
Cryo-Em Structure Of Thehydroxycarboxylic Acid Receptor 2-Gi Protein Complex Bound With Gsk256073
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: MEMBRANE PROTEIN Ligands: OKL |
 |
Organism: Yellow fever virus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2022-11-02 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Yellow fever virus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2022-11-02 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Yellow fever virus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2022-11-02 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-10-26 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome-related coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-10-26 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: MEMBRANE PROTEIN Ligands: Y01, ADP, MG, ATP |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: MEMBRANE PROTEIN Ligands: Y01, ADP, MG, ATP, ESV |

