Search Count: 24
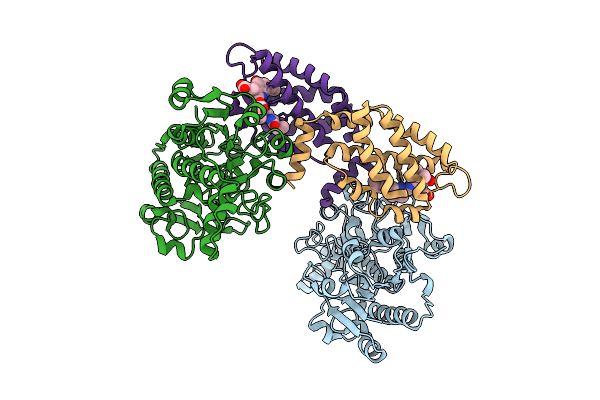 |
Structure Of The Recombinant Structure Of The Subunit Of Allophycocyanin (Apc) And The Formate Dehydrogenase (Fdh)
Organism: Candida boidinii, Picosynechococcus sp. pcc 7003
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: PHOTOSYNTHESIS Ligands: CYC |
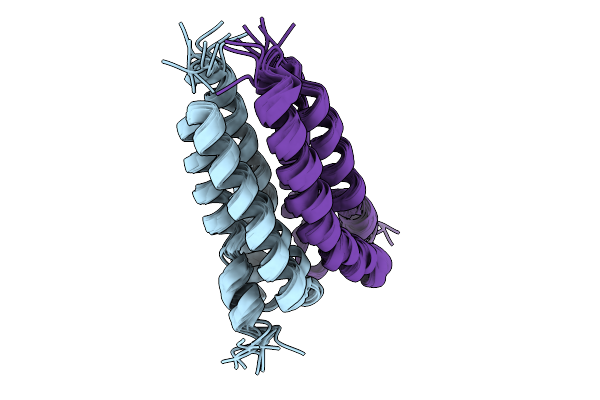 |
The Outward-Open Structure Of Bjsemisweet In Native Cellular Membranes Determined By In Situ Solid-State Nmr
Organism: Bradyrhizobium diazoefficiens usda 110
Method: SOLID-STATE NMR Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN |
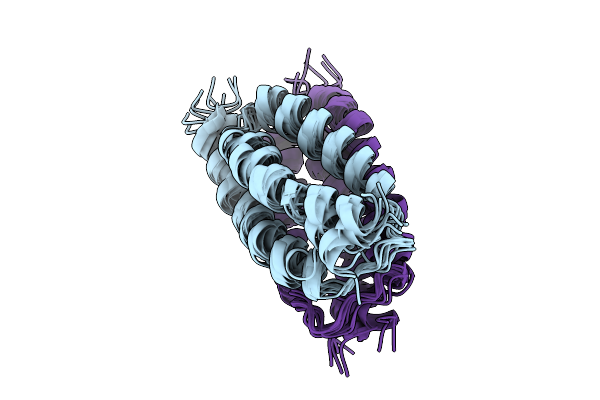 |
The Occluded Structures Of Bjsemisweet In Native Cellular Membranes Determined By In Situ Solid-State Nmr
Organism: Bradyrhizobium diazoefficiens usda 110
Method: SOLID-STATE NMR Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN |
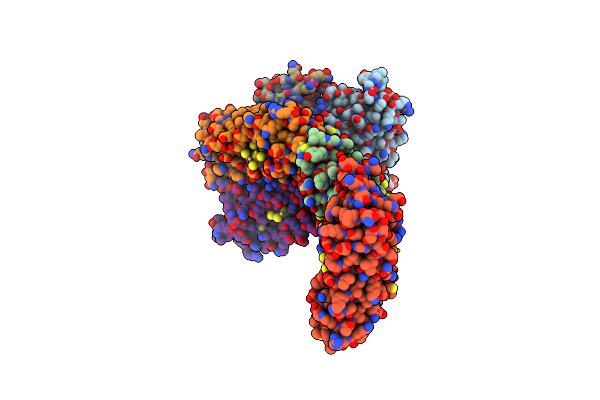 |
Organism: Penaeus japonicus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-05-28 Classification: METAL BINDING PROTEIN Ligands: CD, H2S, MG, SO4 |
 |
Structural Organization Of The Palisade Layer In Isolated Vaccinia Virus Cores
Organism: Vaccinia virus (strain western reserve)
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: VIRAL PROTEIN |
 |
Structural Organization Of The Palisade Layer In Intracellular Mature Vaccinia Virions
Organism: Vaccinia virus (strain western reserve)
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: VIRAL PROTEIN |
 |
Organism: Aetokthonos hydrillicola
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE Ligands: 67I, MLT |
 |
Crystal Structure Of Nitrile Synthase Aetd With Substrate Bound And Cofactor Partially Assembled
Organism: Aetokthonos hydrillicola
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE Ligands: 67I, MLT, SIN, FE, GOL |
 |
Crystal Structure Of Nitrile Synthase Aetd With Substrate Bound And Cofactor Fully Assembled
Organism: Aetokthonos hydrillicola
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE Ligands: 67I, FE |
 |
Cryo-Em Structure Of Sars-Cov-2 Omicron Ba.2 S-Trimer In Complex With Fab L4.65 And L5.34
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Omicron Ba.4 S-Trimer In Complex With Fab L4.65 And L5.34
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Omicron Prototype S-Trimer In Complex With Fab L4.65 And L5.34
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: IMMUNE SYSTEM/VIRAL PROTEIN |
 |
Cryo-Em Structure Of Sars-Cov-2 Omicron Ba.2 Rbd In Complex With Fab L4.65 And L5.34
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Omicron Ba.4 Rbd In Complex With Fab L4.65 And L5.34
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Omicron Prototype Rbd In Complex With Fab L4.65 And L5.34
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: IMMUNE SYSTEM/VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Ba.2 Rbd In Complex With Ba7535 Fab (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 Delta Spike Protein In Complex With Ba7208 And Ba7125 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Delta Rbd In Complex With Ba7054 And Ba7125 Fab (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Sars-Cov-2 Delta Rbd In Complex With Ba7208 And Ba7125 Fab (Local Refinement)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-03-01 Classification: VIRAL PROTEIN |

