Search Count: 17
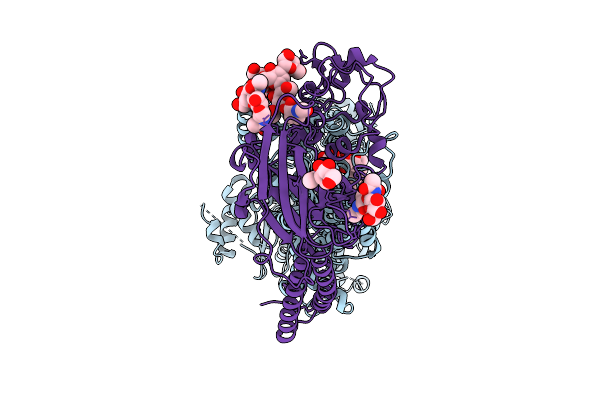 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
 |
Cryo-Em Structure Of The S. Cerevisiae Lipid Flippase Neo1 Bound With Pi4P In The E2P State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: LIPID TRANSPORT Ligands: BEF, A1ARJ |
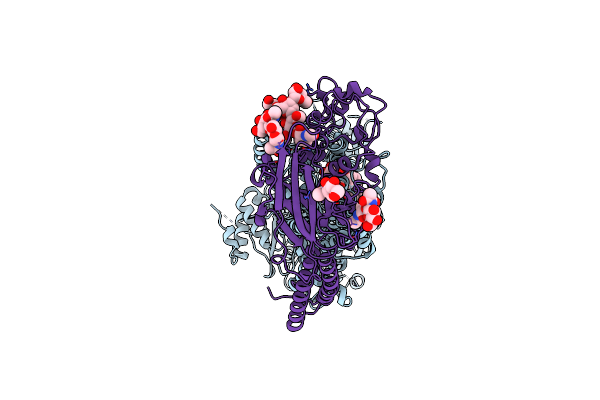 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
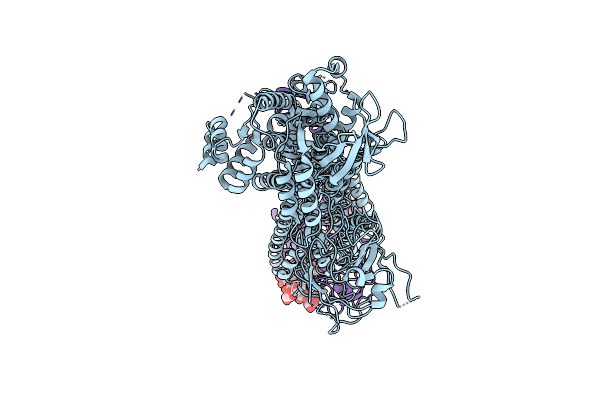 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 In The E1 State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: LIPID TRANSPORT Ligands: NAG, MG |
 |
Cryo-Em Structure Of The S. Cerevisiae Arf-Like Protein Arl1 Bound To The Arf Guanine Nucleotide Exchange Factor Gea2
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: LIPID TRANSPORT Ligands: GTP |
 |
Cryo-Em Structure Of The S. Cerevisiae Guanine Nucleotide Exchange Factor Gea2
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: LIPID TRANSPORT |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: TRANSLOCASE Ligands: BEF, MG |
 |
Structure Of The S. Cerevisiae P4B Atpase Lipid Flippase In The E2P-Transition State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: TRANSLOCASE Ligands: ALF, MG |
 |
Structure Of The S. Cerevisiae P4B Atpase Lipid Flippase In The E1-Atp State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: TRANSLOCASE Ligands: ACP, MG |
 |
Structure Of The S. Cerevisiae Phosphatidylcholine Flippase Dnf2-Lem3 Complex In The E2P Transition State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: TRANSLOCASE Ligands: CLR, ALF, MG, NAG |
 |
Structure Of The S. Cerevisiae Phosphatidylcholine Flippase Dnf1-Lem3 Complex In The Apo E1 State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: TRANSLOCASE Ligands: MG, NAG |
 |
Structure Of The S. Cerevisiae Phosphatidylcholine Flippase Dnf2-Lem3 Complex In The Apo E1 State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: TRANSLOCASE Ligands: MG, CLR |
 |
Structure Of The S. Cerevisiae Phosphatidylcholine Flippase Dnf2-Lem3 Complex In The E1-Atp State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: TRANSLOCASE Ligands: CLR, MG, ACP, MAN, NAG |
 |
Structure Of The S. Cerevisiae Phosphatidylcholine Flippase Dnf2-Lem3 Complex In The E1-Adp State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: TRANSLOCASE Ligands: ADP, ALF, MG, NAG, MAN |
 |
Structure Of The S. Cerevisiae Phosphatidylcholine Flippase Dnf2-Lem3 Complex In The E2P State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: TRANSLOCASE Ligands: CLR, MG, BEF, NAG |
 |
Structure Of The S. Cerevisiae Phosphatidylcholine Flippase Dnf1-Lem3 Complex In The E1-Adp State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: TRANSLOCASE Ligands: POV, ADP, ALF, MG, NAG |
 |
Structure Of The S. Cerevisiae Phosphatidylcholine Flippase Dnf1-Lem3 Complex In The E2P State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: TRANSLOCASE Ligands: BEF, MG, POV, NAG |

