Search Count: 25
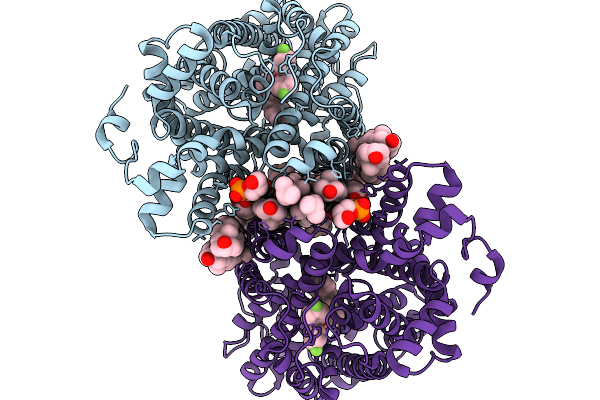 |
Cryo-Em Structure Of Lipid-Mediated Dimer Of Human Norepinephrine Transporter Net In The Presence Of Vanoxerine In An Inward-Open State At Resolution Of 2.52 Angstrom
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: CLR, A1D5S, A1EFM |
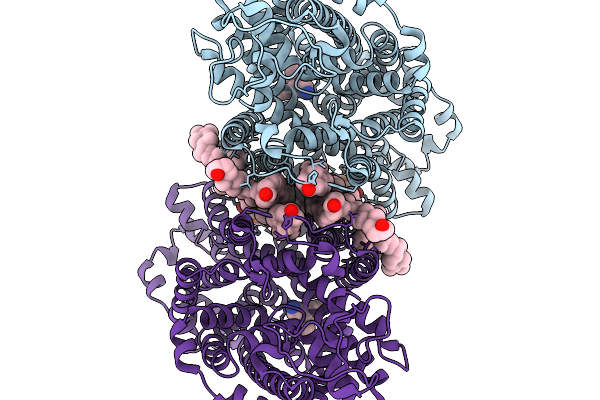 |
Cryo-Em Structure Of Lipid-Mediated Human Norepinephrine Transporter Net In The Presence Of Levomilnacipran In An Inward-Open State At Resolution Of 2.46 Angstrom.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: F0F, CLR, A1EFM |
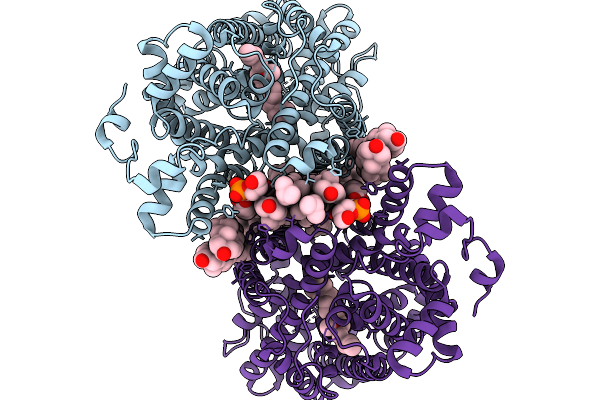 |
Cryo-Em Structure Of Lipid-Mediated Dimer Of Human Norepinephrine Transporter Net In The Presence Of The F3288-0031 In An Inward-Open State At Resolution Of 3.1 Angstrom
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: A1EFR, CLR, A1EFM |
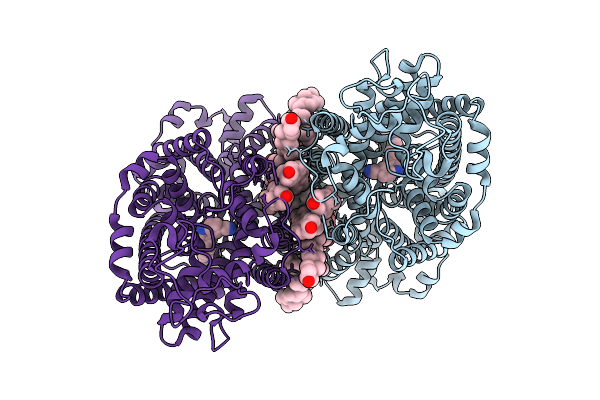 |
Cryo-Em Structure Of Lipid-Mediated Dimer Of Human Norepinephrine Transporter Net In The Presence Of The Antidepressant Vilazodone In An Inward-Open State At Resolution Of 2.44 Angstrom.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: PROTON TRANSPORT Ligands: YG7, CLR, A1EFM |
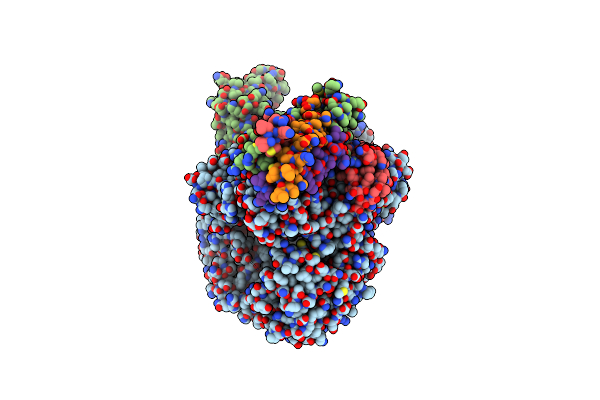 |
Cryo-Em Structure Of The Rna-Dependent Rna Polymerase Complex From Marburg Virus
Organism: Marburg virus - musoke, kenya, 1980, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2025-04-09 Classification: VIRAL PROTEIN Ligands: ZN |
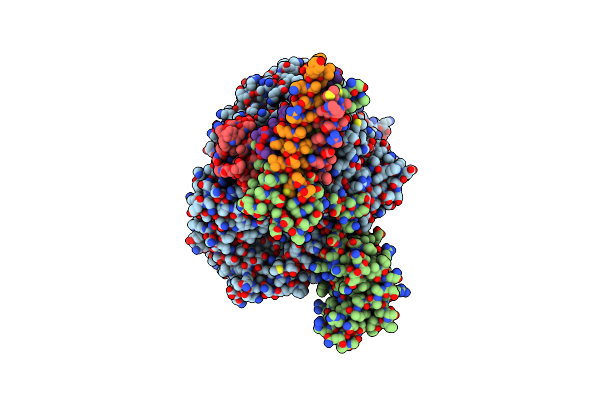 |
Cryo-Em Structure Of The Rna-Dependent Rna Polymerase Complex In A Compact Conformation From Ebola Virus
Organism: Escherichia coli (strain k12), Ebola virus - eckron (zaire, 1976)
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-04-09 Classification: VIRAL PROTEIN Ligands: ZN |
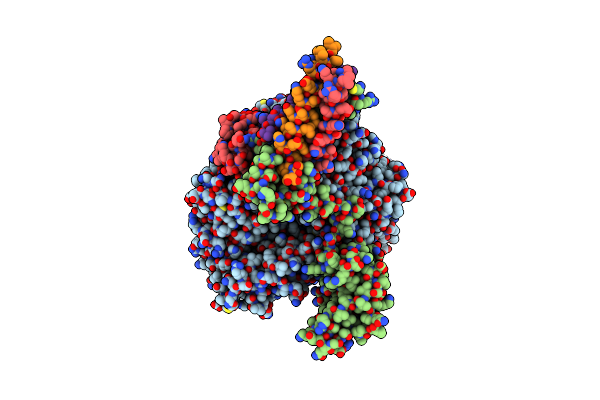 |
Cryo-Em Structure Of The Rna-Dependent Rna Polymerase Complex From Marburg Virus
Organism: Marburg virus - musoke, kenya, 1980, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:2.84 Å Release Date: 2025-04-09 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: HYDROLASE Ligands: A1A4Y, ZN, A1BHF |
 |
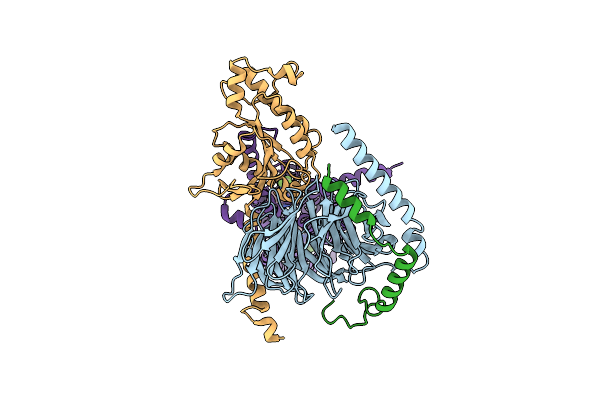
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: MEMBRANE PROTEIN Ligands: A1AEI |
 |
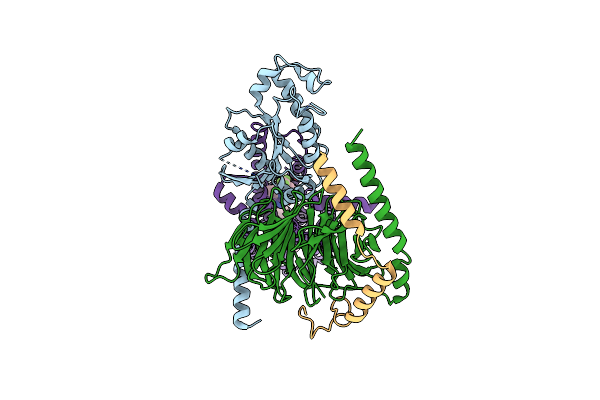
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: MEMBRANE PROTEIN Ligands: A1AEI |
 |
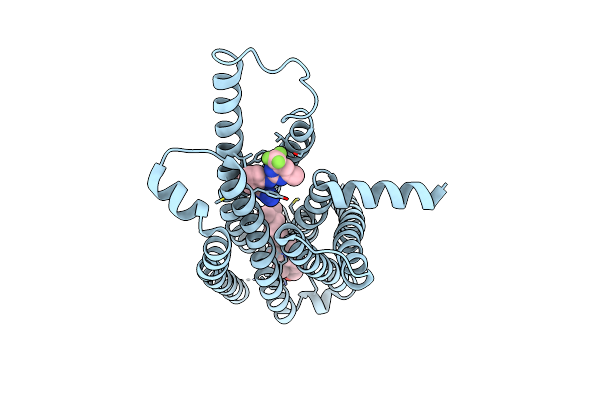
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: MEMBRANE PROTEIN Ligands: A1AEI, CLR |
 |
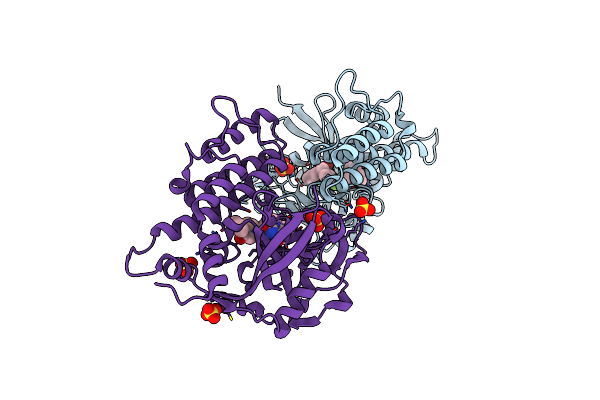
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: MEMBRANE PROTEIN Ligands: A1AEI, CLR |
 |
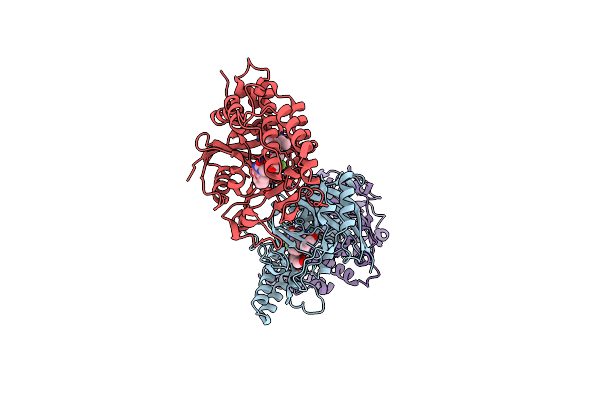
Adp-Ribosyltransferase 1 (Parp1) Catalytic Domain Bound To A Quinazoline-2,4(1H,3H)-Dione Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-11-08 Classification: TRANSFERASE Ligands: 1WI, SO4 |
 |
Human Adp-Ribosyltransferase 2 (Parp2) Catalytic Domain Bound To A Quinazoline-2,4(1H,3H)-Dione Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2023-11-08 Classification: TRANSFERASE Ligands: GOL, 1WI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2023-10-11 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: V4I |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-10-11 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: V59 |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-07-26 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, GDP, MES, IUK |
 |
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-12-14 Classification: ELECTRON TRANSPORT Ligands: PO4 |
 |
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-12-14 Classification: ELECTRON TRANSPORT Ligands: MG, CL |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With To-317
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-06-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: TO3, EDO, FMT, ZN, K |

