Search Count: 485
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: CA, YUY, YUV, A1CGW |
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: NAG, CA, YUV, A1CGW, YUY |
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: NAG, YUY, YUV, A1CGW |
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: NAG, CA |
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: NAG, CA |
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: NAG, CA, A1CGZ, A1CGW, A1CG0 |
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: NAG, CA, YUY, YUV, A1CG5 |
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSFERASE Ligands: NAG, CA, A1CGZ, A1CGW, A1CG0 |
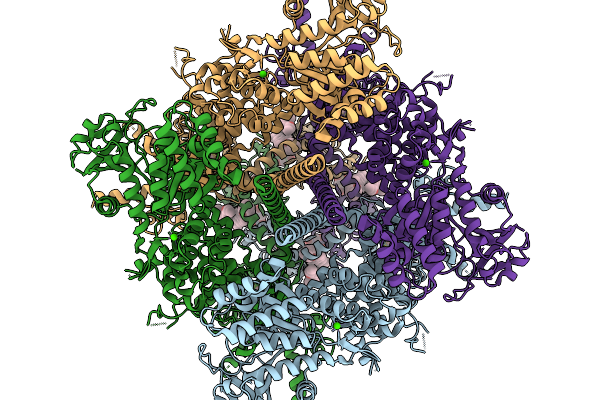 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSFERASE Ligands: NAG, CA, A1CGZ, A1CGW, A1CG0 |
 |
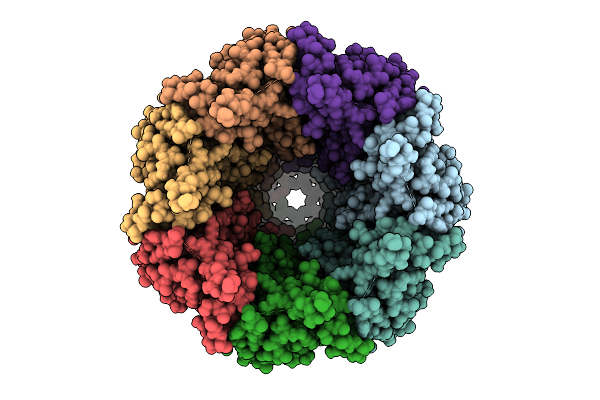
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSFERASE Ligands: NAG, CA, A1CGZ, A1CG0 |
 |
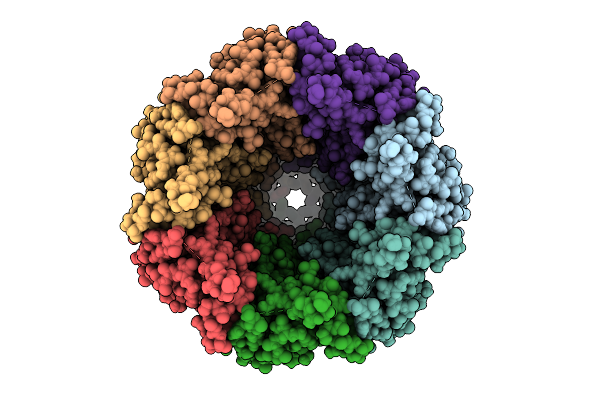
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: PTY, CLR, NAG |
 |
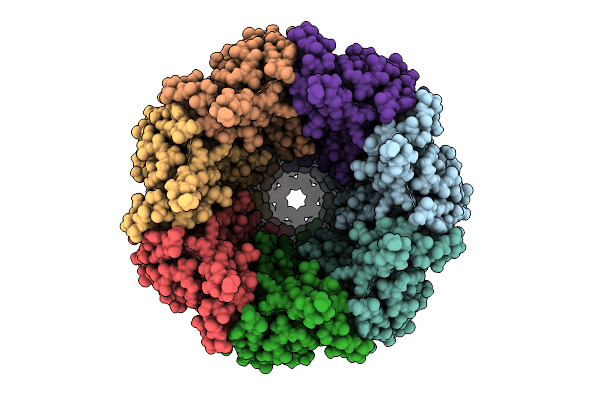
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: PTY, CLR, NAG |
 |
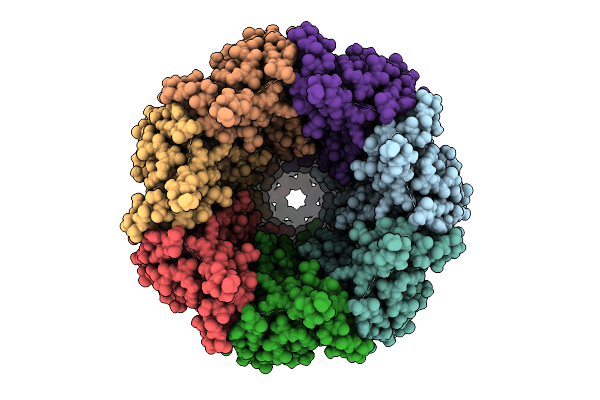
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: PTY, CLR, NAG |
 |
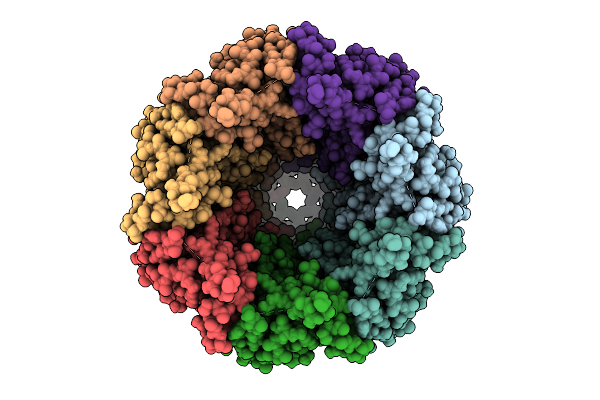
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: PTY, CLR, NAG |
 |
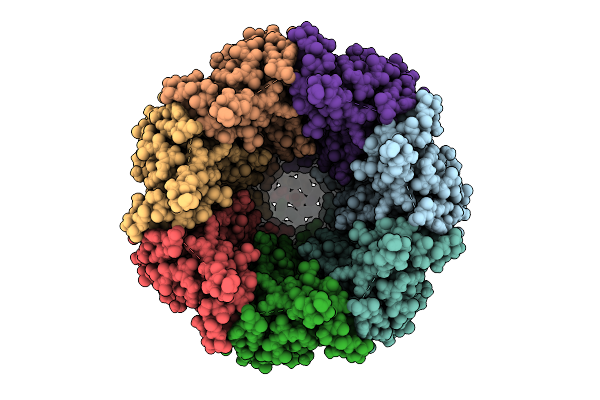
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: PTY, CLR, NAG |
 |
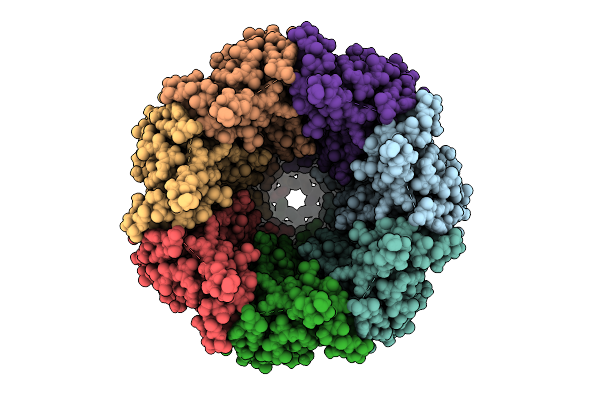
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: PTY, CLR, NAG, ATP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: PTY, CLR, NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: PTY, CLR, NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: PTY, CLR, NAG |
 |
Organism: Homo sapiens, Human respiratory syncytial virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: A1A1V |

