Search Count: 44
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: STRUCTURAL PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: STRUCTURAL PROTEIN |
 |
Organism: Autographa californica nucleopolyhedrovirus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: STRUCTURAL PROTEIN Ligands: NAG |
 |
Organism: Autographa californica nucleopolyhedrovirus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: STRUCTURAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-02-22 Classification: CELL INVASION Ligands: GDP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-02-22 Classification: CELL INVASION Ligands: GOL, GDP, MG |
 |
Organism: Penicillium rubens wisconsin 54-1255
Method: SOLUTION NMR Release Date: 2022-08-24 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Penicillium rubens (strain atcc 28089 / dsm 1075 / nrrl 1951 / wisconsin 54-1255)
Method: SOLUTION NMR Release Date: 2022-03-30 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-02-16 Classification: ISOMERASE Ligands: J2C, PE8 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2022-02-16 Classification: ISOMERASE Ligands: J3X |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-02-16 Classification: ISOMERASE Ligands: J50, PE8 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-02-16 Classification: ISOMERASE Ligands: 0BF, P33 |
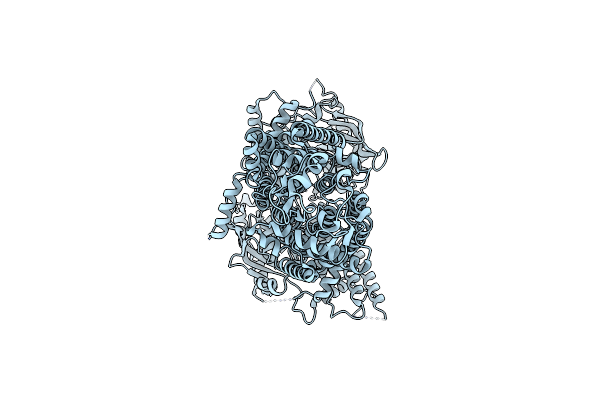 |
Cryo-Em Structure Of Pdr5 From Saccharomyces Cerevisiae In Inward-Facing Conformation Without Nucleotides
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: TRANSPORT PROTEIN |
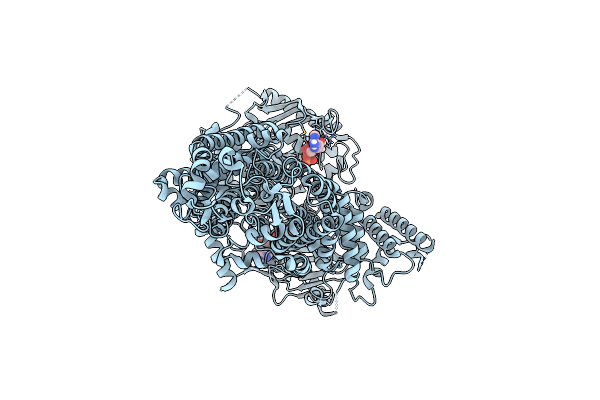 |
Cryo-Em Structure Of Pdr5 From Saccharomyces Cerevisiae In Inward-Facing Conformation With Adp/Atp
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: TRANSPORT PROTEIN Ligands: ATP, ADP |
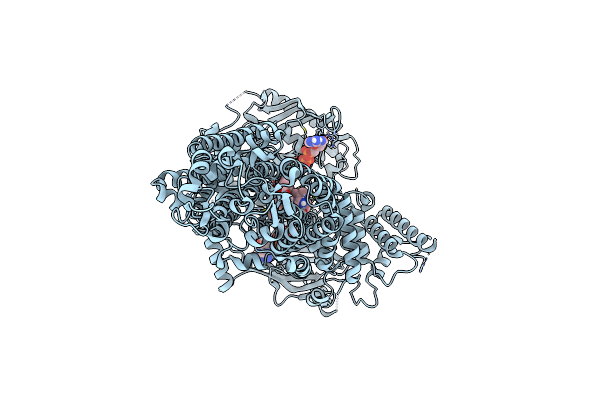 |
Cryo-Em Structure Of Pdr5 From Saccharomyces Cerevisiae In Inward-Facing Conformation With Adp/Atp And Rhodamine 6G
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: TRANSPORT PROTEIN Ligands: ATP, ADP, RHQ |
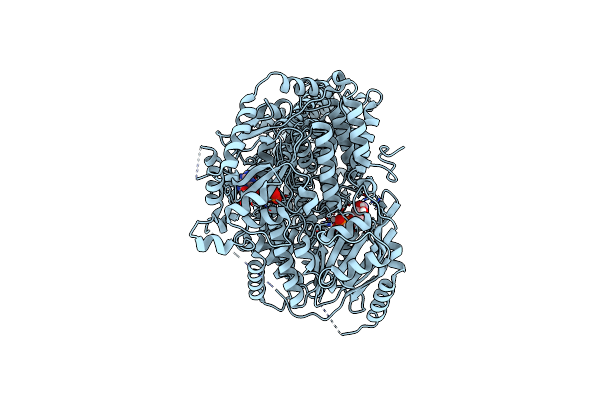 |
Cryo-Em Structure Of Pdr5 From Saccharomyces Cerevisiae In Outward-Facing Conformation With Adp-Orthovanadate/Atp
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, AOV |
 |
Structure Of The Human Glun1-Glun2A Nmda Receptor In Complex With S-Ketamine, Glycine And Glutamate
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: MEMBRANE PROTEIN Ligands: NAG, GLY, GLU, JC9 |
 |
Structure Of The Human Glun1-Glun2B Nmda Receptor In Complex With S-Ketamine,Glycine And Glutamate
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-07-28 Classification: MEMBRANE PROTEIN Ligands: NAG, JC9 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-02-24 Classification: GENE REGULATION Ligands: XB7, UNX |
 |
Cryo-Em Structure Of Escherichia Coli Acrbz And Darpin In Saposin A-Nanodisc With Cardiolipin
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: MEMBRANE PROTEIN |

