Search Count: 42
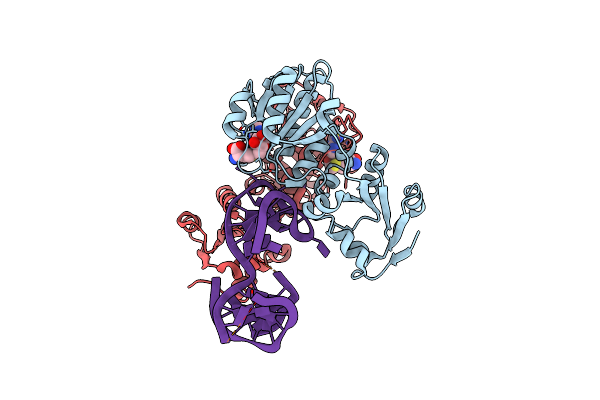 |
Rlmr 23S Rrna Methyltransferase From Thermus Thermophilus In Complex With Rrna And S-Adenosyl-L-Homocysteine (Sah)
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: TRANSFERASE Ligands: SAH |
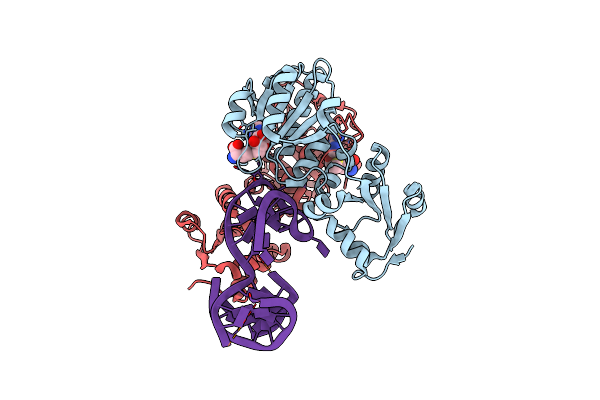 |
Rlmr 23S Rrna Methyltransferase From Thermus Thermophilus In Complex With Methylated Rrna (Um2552) And S-Adenosyl-L-Homocysteine (Sah)
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: TRANSFERASE/RNA Ligands: SAH, SAM |
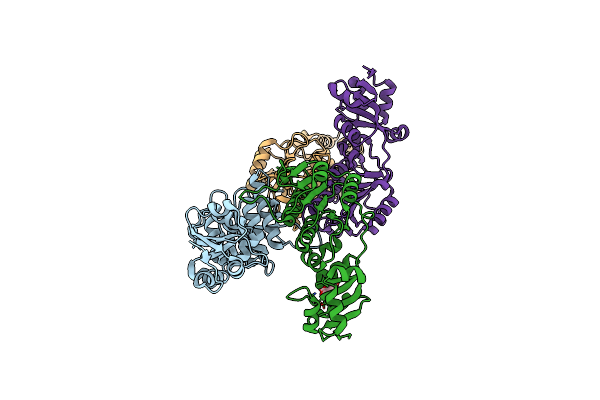 |
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: TRANSFERASE Ligands: CIT |
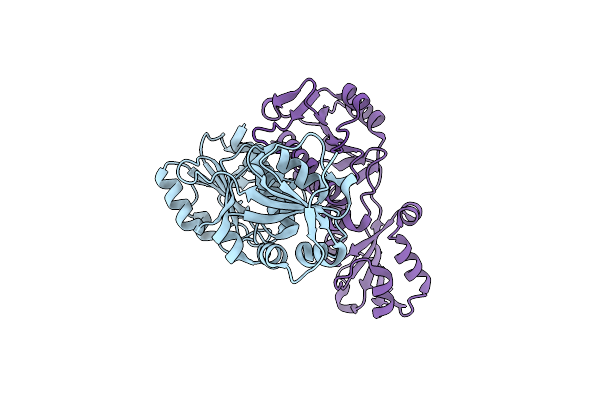 |
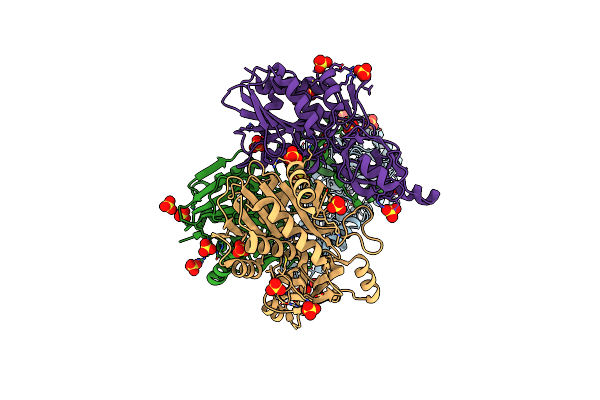
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-12-28 Classification: TRANSFERASE |
 |
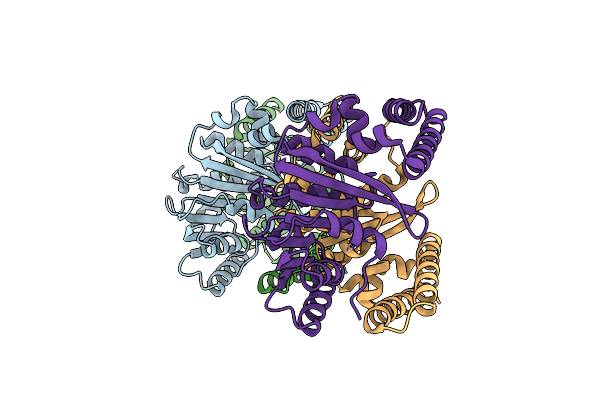
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-10-23 Classification: HYDROLASE Ligands: ZN, CO, SO4, OH |
 |
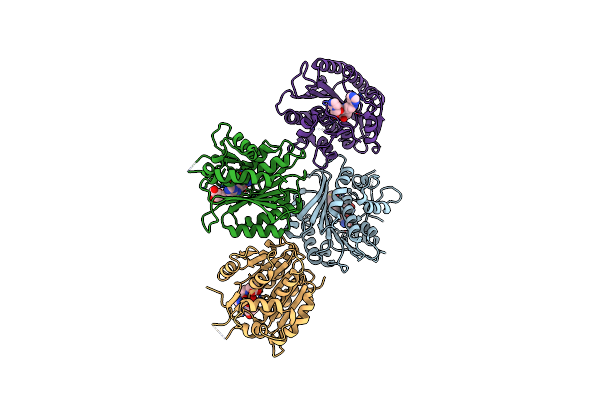
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-03-27 Classification: RNA BINDING PROTEIN Ligands: NI |
 |
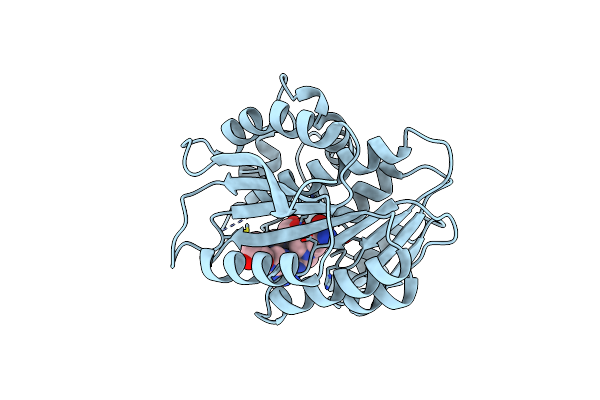
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-03-27 Classification: TRANSFERASE Ligands: HY8 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2019-03-27 Classification: TRANSFERASE Ligands: HZ2 |
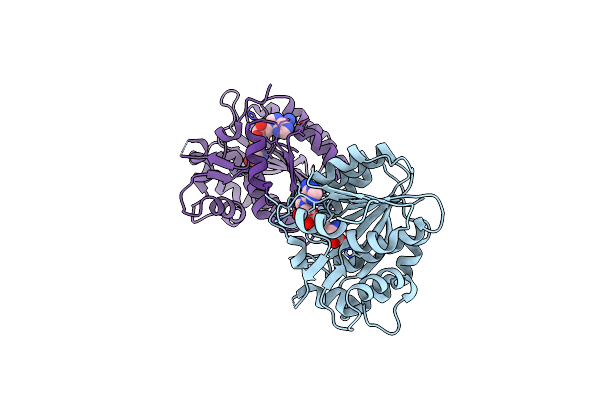 |
Structure Of E.Coli Rlmj In Complex With The Natural Cofactor Product S-Adenosyl-Homocysteine (Sah)
Organism: Escherichia coli pcn033
Method: X-RAY DIFFRACTION Release Date: 2019-03-27 Classification: TRANSFERASE Ligands: SAH |
 |
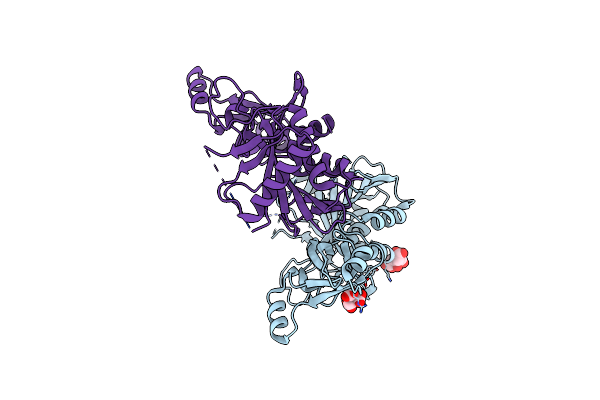
Structure Of M. Capricolum Trmk In Complex With The Natural Cofactor Product S-Adenosyl-Homocysteine (Sah)
Organism: Mycoplasma capricolum subsp. capricolum
Method: X-RAY DIFFRACTION Release Date: 2019-03-27 Classification: TRANSFERASE Ligands: SAH |
 |
Crystal Structure Of Dual Specific Trm10 Construct From Thermococcus Kodakaraensis.
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-06-13 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of Dual Specific Trm10 Construct From Thermococcus Kodakaraensis.
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-06-13 Classification: RNA BINDING PROTEIN Ligands: ACT, GOL, TRS |
 |
Crystal Structure Of Dual Specific Trm10 Construct From Thermococcus Kodakaraensis.
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-06-13 Classification: RNA BINDING PROTEIN Ligands: SAM, GOL |
 |
Crystal Structure Of Dual Specific Trm10 Construct From Thermococcus Kodakaraensis.
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Release Date: 2018-06-13 Classification: RNA BINDING PROTEIN Ligands: SAH |
 |
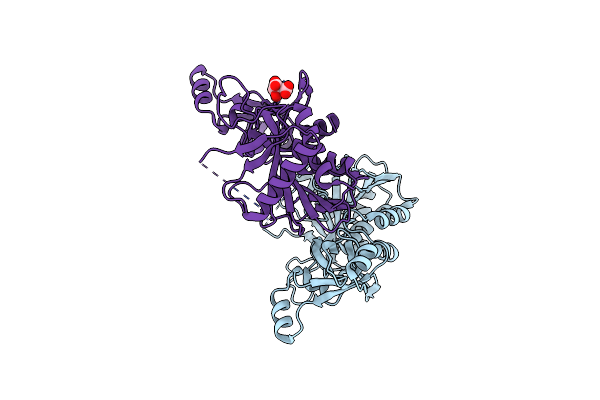
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-05-16 Classification: LYASE Ligands: CIT |
 |
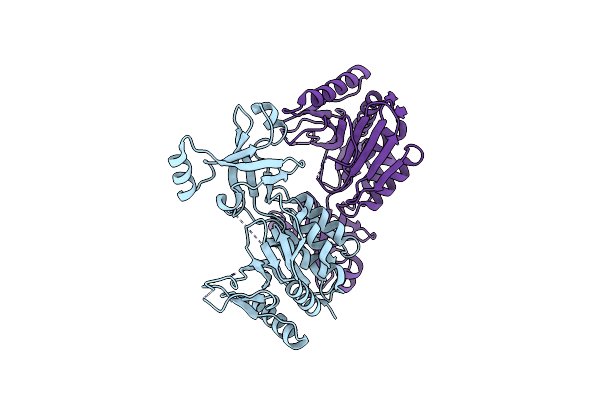
Crystal Structure Of H60A Mutant Of Thermotoga Maritima Tmpep1050 Aminopeptidase
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2018-05-16 Classification: LYASE Ligands: CIT |
 |
Crystal Structure Of H307A Mutant Of Thermotoga Maritima Tmpep1050 Aminopeptidase
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-05-16 Classification: LYASE |
 |
Crystal Structure Of H60A H307A Mutant Of Thermotoga Maritima Tmpep1050 Aminopeptidase
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2018-05-16 Classification: LYASE |
 |
Crystal Structure Of D62A Mutant Of Thermotoga Maritima Tmpep1050 Aminopeptidase
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-06-21 Classification: HYDROLASE Ligands: CIT, NA |
 |
Crystal Structure Of Sulfolobus Acidocaldarius Trm10 At 2.4 Angstrom Resolution.
Organism: Sulfolobus acidocaldarius
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-01-13 Classification: TRANSFERASE Ligands: PEG |

