Search Count: 24
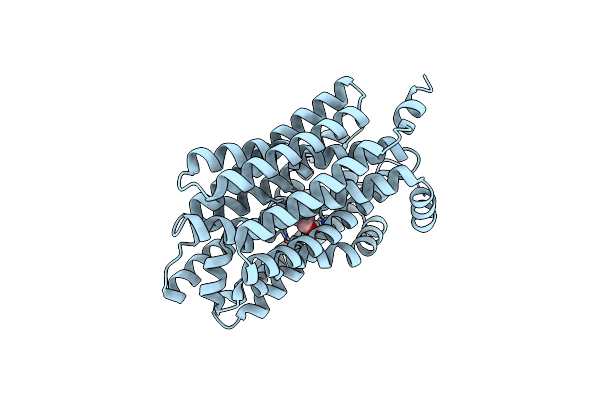 |
Cryo-Em Structure Of The Inward-Open Choline-Bound State Of Choline/Ethanolamine Transporter Flvcr2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN Ligands: CHT |
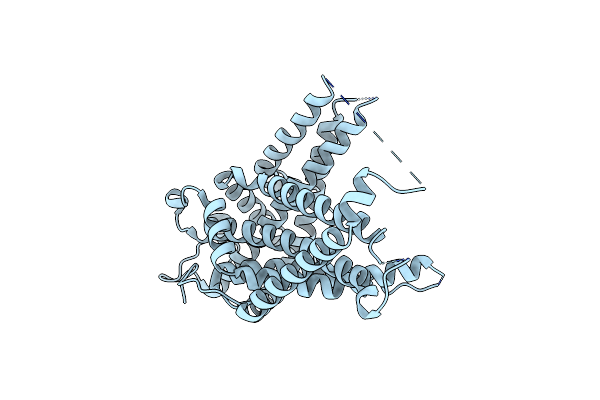 |
Cryo-Em Structure Of The Human Choline Transporter-Like Protein Hctl1 In Lmng
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN |
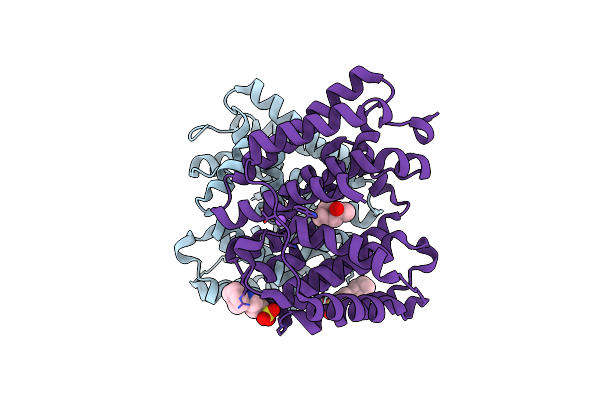 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-24 Classification: LYASE Ligands: CXS, NA, MPD |
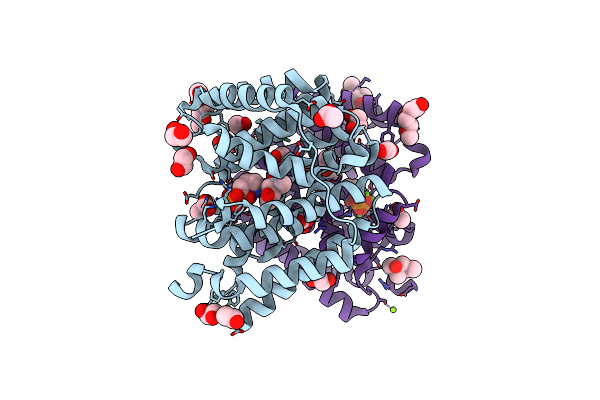 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-04-24 Classification: LYASE Ligands: AHD, MG, EDO, MPD, ACT, PG0, CL |
 |
Crystal Structure Of A Selenomethionine-Labeled Hydropyrene Synthase (M75L Variant) In Its Closed Conformation
Organism: Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:3.39 Å Release Date: 2023-10-04 Classification: LYASE Ligands: MG, AHD |
 |
Crystal Structure Of A Hydropyrene Synthase (M75L Variant) In Its Closed Conformation
Organism: Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2023-10-04 Classification: LYASE Ligands: MG, 212 |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-12-02 Classification: LYASE Ligands: AHD, MPD, MG, CL |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-02 Classification: LYASE Ligands: AHD, MG, CL, MPD |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-12-02 Classification: LYASE Ligands: MPD, AHD, MG, CL |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-12-02 Classification: LYASE Ligands: AHD, MPD, MG, CL |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-12-02 Classification: LYASE Ligands: MPD |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2020-12-02 Classification: LYASE Ligands: MPD, MG |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-10-10 Classification: LYASE Ligands: MG, PPV, EXW, MPD, CL, CCN, PEG, K |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-10-10 Classification: LYASE Ligands: AHD, MG |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-10-10 Classification: LYASE Ligands: MG, CL |
 |
Organism: Streptomyces melanosporofaciens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-10-10 Classification: LYASE Ligands: MG, CL |
 |
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-08-15 Classification: PROTEIN BINDING Ligands: BWH, SO4 |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2018-01-24 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, PG4, HEM |
 |
Organism: Rhodococcus erythropolis dn1
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2017-11-29 Classification: LYASE Ligands: GOL, FMT, MG |
 |
Crystal Structure Of Hla-B*27:05 Complexed With The Self-Peptide Pvipr Measured At 295 K
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2017-02-01 Classification: IMMUNE SYSTEM |

