Search Count: 56
 |
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-07-13 Classification: PLANT PROTEIN |
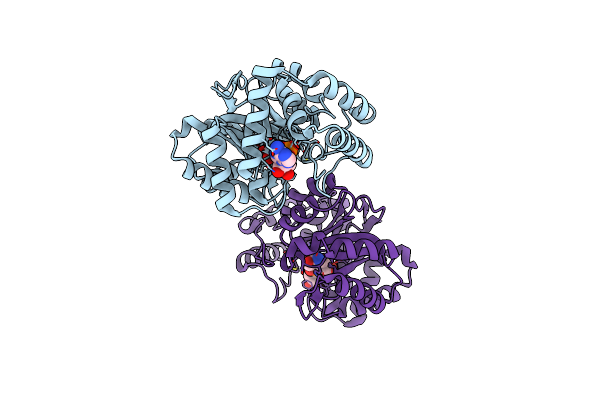 |
Crystal Structure Of L-Galactose Dehydrogenase From Spinacia Oleracea In Complex With Nad+
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-07-13 Classification: PLANT PROTEIN Ligands: NAD |
 |
Organism: Fusobacterium nucleatum subsp. nucleatum atcc 25586
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-03-24 Classification: HYDROLASE Ligands: GOL, MG, SPD |
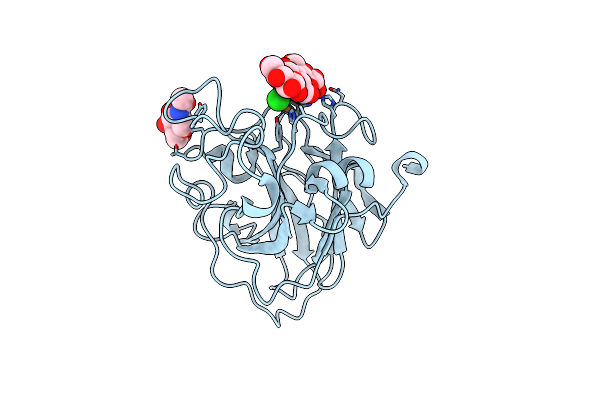 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-03-02 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |
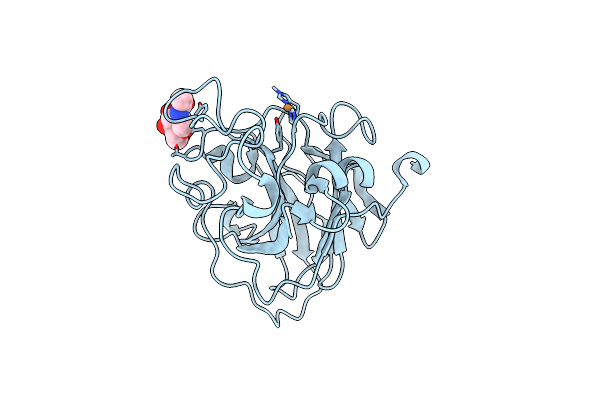 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2016-03-02 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |
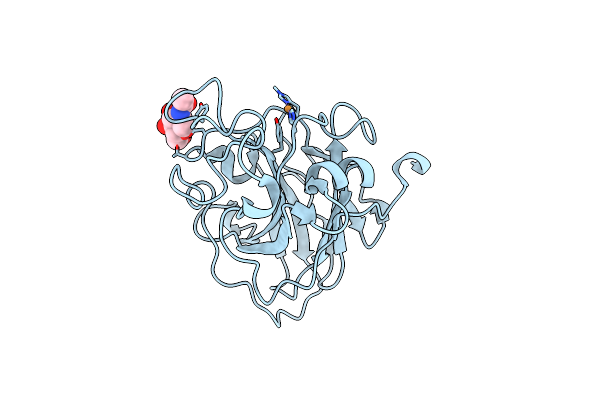 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2016-03-02 Classification: OXIDOREDUCTASE Ligands: CL, CU, NAG |
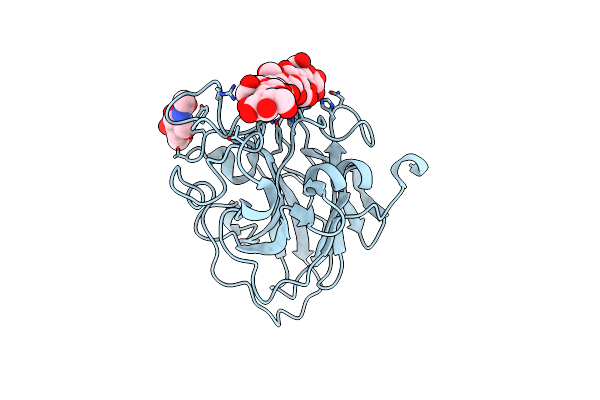 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-03-02 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |
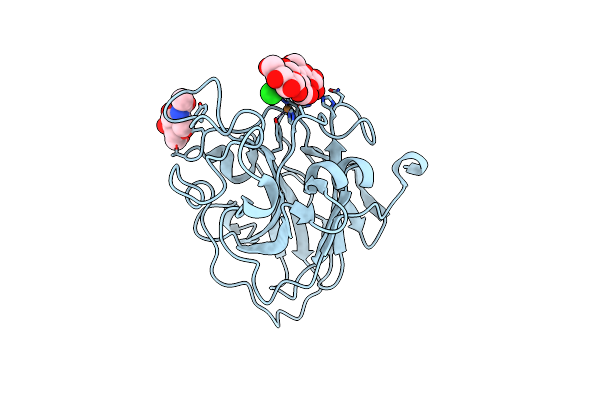 |
Organism: Lentinus similis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-03-02 Classification: OXIDOREDUCTASE Ligands: CU, NAG, CL |
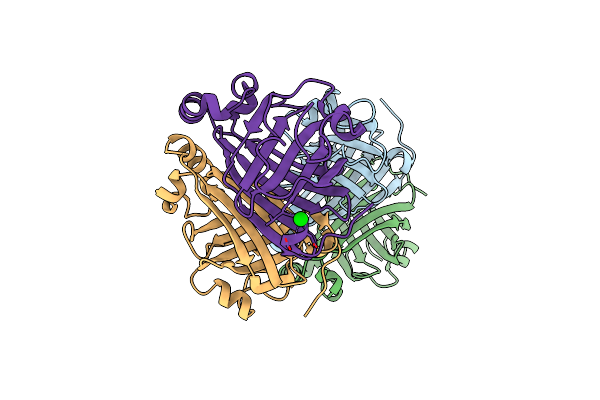 |
Crystal Structure Of Single Point Mutant Arg48Gln Of P-Coumaric Acid Decarboxylase From Lactobacillus Plantarum Structural Insights Into The Active Site And Decarboxylation Catalytic Mechanism
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2010-02-09 Classification: LYASE Ligands: BA |
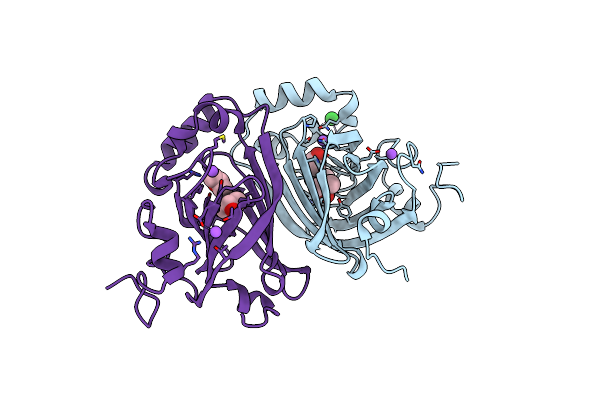 |
Crystal Structure Of Single Point Mutant Glu71Ser P-Coumaric Acid Decarboxylase
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2010-02-09 Classification: LYASE Ligands: BA, IPA, NA, CL |
 |
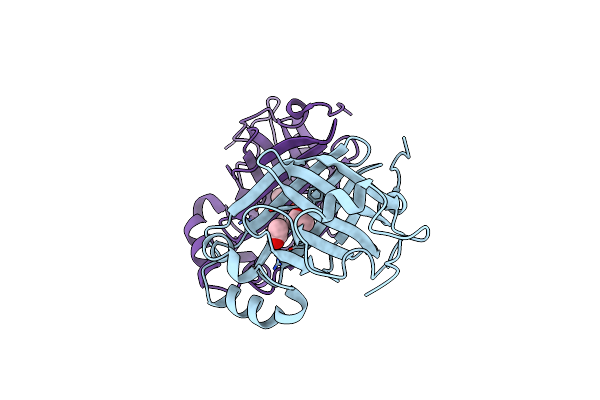
Crystal Structure Of P-Coumaric Acid Decarboxylase From Lactobacillus Plantarum: Structural Insights Into The Active Site And Decarboxylation Catalytic Mechanism
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2009-11-17 Classification: LYASE |
 |
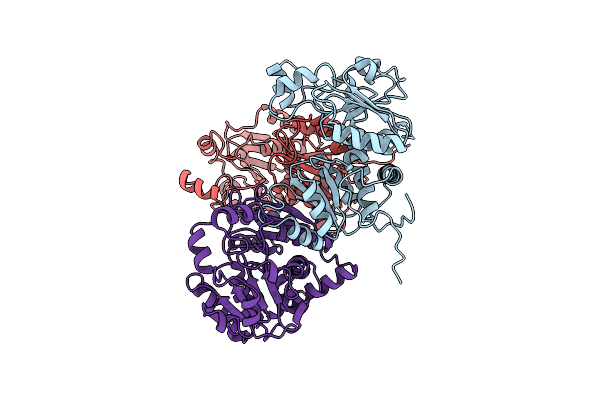
Crystal Structure Of Single Point Mutant Tyr20Phe P-Coumaric Acid Decarboxylase From Lactobacillus Plantarum: Structural Insights Into The Active Site And Decarboxylation Catalytic Mechanism
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2009-11-17 Classification: LYASE Ligands: ACT, IPA |
 |
Crystal Structure Of The Hexameric Catabolic Ornithine Transcarbamylase From Lactobacillus Hilgardii
Organism: Lactobacillus hilgardii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-11-17 Classification: TRANSFERASE Ligands: NI |
 |
Crystal Structure Of The Mutant E55Q Of The Cellulase Cel48F In Complex With A Thio-Oligosaccharide
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-01-15 Classification: HYDROLASE Ligands: CA |
 |
Catalytic Domain Of Endo-1,4-Glucanase Cel6A From Thermobifida Fusca In Complex With Methyl Cellobiosyl-4-Thio-Beta-Cellobioside
Organism: Thermomonospora fusca
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2005-10-05 Classification: HYDROLASE |
 |
Catalytic Domain Of Endo-1,4-Glucanase Cel6A Mutant Y73S From Thermobifida Fusca
Organism: Thermomonospora fusca
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2005-10-05 Classification: HYDROLASE |
 |
Catalytic Domain Of Endo-1,4-Glucanase Cel6A Mutant Y73S From Thermobifida Fusca In Complex With Cellotetrose
Organism: Thermomonospora fusca
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2005-10-05 Classification: HYDROLASE |
 |
Catalytic Domain Of Endo-1,4-Glucanase Cel6A Mutant Y73S From Thermobifida Fusca In Complex With Methyl Cellobiosyl-4-Thio-Beta- Cellobioside
Organism: Thermomonospora fusca
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2005-10-05 Classification: HYDROLASE |
 |
Cellobiohydrolase Cel7A With Disulphide Bridge Added Across Exo-Loop By Mutations D241C And D249C
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2003-11-25 Classification: HYDROLASE Ligands: NAG, CO |
 |
Cellobiohydrolase Cel7A With Loop Deletion 245-252 And Bound Non-Hydrolysable Cellotetraose
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2003-11-25 Classification: HYDROLASE Ligands: NAG, CA |

