Search Count: 34
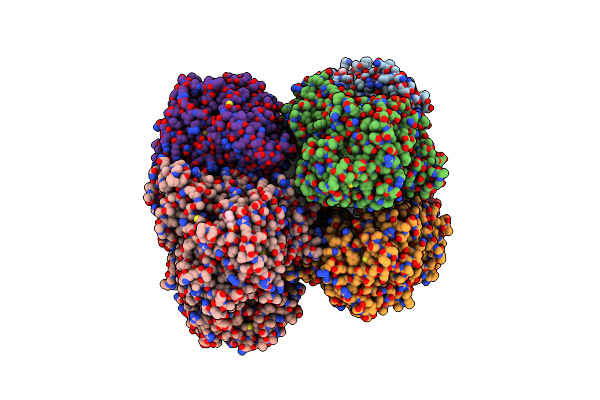 |
The Structure Of Aerobactin Synthetase Iucc From A Hypervirulent Pathotype Of Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-04-11 Classification: LIGASE Ligands: MES |
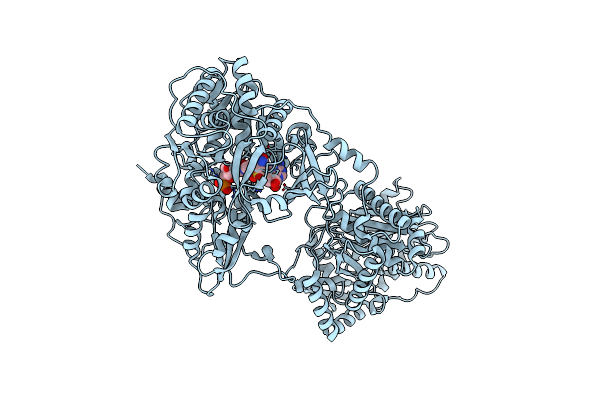 |
Crystal Structure Of Holo-Entf A Nonribosomal Peptide Synthetase In The Thioester-Forming Conformation
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-09-21 Classification: BIOSYNTHETIC PROTEIN Ligands: 75C |
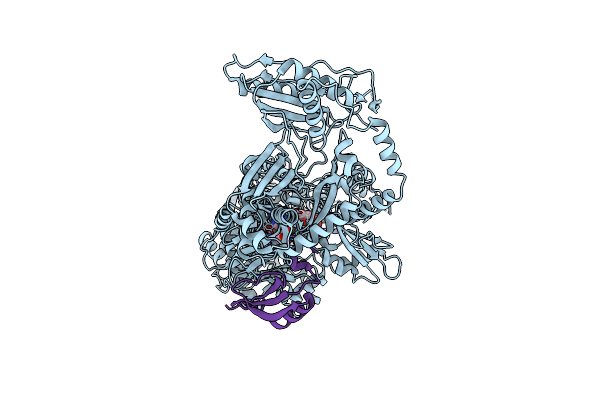 |
Entf, A Terminal Nonribosomal Peptide Synthetase Module Bound To The Mbth-Like Protein Ybdz
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-09-14 Classification: LIGASE Ligands: 75C, CL |
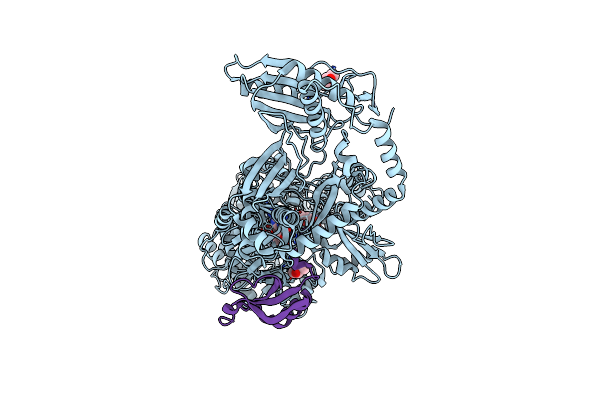 |
Entf, A Terminal Nonribosomal Peptide Synthetase Module Bound To The Non-Native Mbth-Like Protein Pa2412
Organism: Escherichia coli (strain k12), Pseudomonas aeruginosa (strain atcc 15692 / pao1 / 1c / prs 101 / lmg 12228)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-09-14 Classification: LIGASE Ligands: 75C, BU9 |
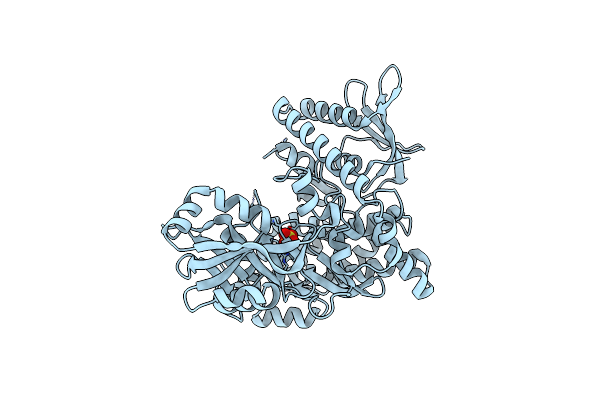 |
The Structure Of Aerobactin Synthetase Iuca From A Hypervirulent Pathotype Of Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-06-15 Classification: LIGASE Ligands: SO4 |
 |
The Structure Of Atp-Bound Aerobactin Synthetase Iuca From A Hypervirulent Pathotype Of Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-06-15 Classification: LIGASE Ligands: ATP, MG |
 |
Structure-Function Studies On Role Of Hydrophobic Clamping Of A Basic Glutamate In Catalysis By Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2016-05-18 Classification: ISOMERASE |
 |
Structure-Function Studies On Role Of Hydrophobic Clamping Of A Basic Glutamate In Catalysis By Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2016-05-18 Classification: ISOMERASE |
 |
Structure-Function Studies On Role Of Hydrophobic Clamping Of A Basic Glutamate In Catalysis By Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-05-18 Classification: ISOMERASE Ligands: K, PGA |
 |
Structure-Function Studies On Role Of Hydrophobic Clamping Of A Basic Glutamate In Catalysis By Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-05-18 Classification: ISOMERASE Ligands: PGA |
 |
Structure-Function Studies On Role Of Hydrophobic Clamping Of A Basic Glutamate In Catalysis By Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-05-18 Classification: ISOMERASE |
 |
Structure-Function Studies On Role Of Hydrophobic Clamping Of A Basic Glutamate In Catalysis By Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2016-05-18 Classification: ISOMERASE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2016-05-11 Classification: TRANSFERASE Ligands: PLP, EDO |
 |
Crystal Structure Of Holo-Ab3403 A Four Domain Nonribosomal Peptide Synthetase From Acinetobacter Baumanii
Organism: Acinetobacter baumannii (strain ab307-0294)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-12-30 Classification: BIOSYNTHETIC PROTEIN Ligands: PNS, PX4, K, NI |
 |
Crystal Structure Of Holo-Ab3403 A Four Domain Nonribosomal Peptide Synthetase Bound To Amp And Glycine
Organism: Acinetobacter baumannii (strain ab307-0294)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-12-30 Classification: BIOSYNTHETIC PROTEIN Ligands: PNS, GLY, AMP, XP4, NI, MG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2014-06-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: EDO, A08 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-06-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: EDO, 1OQ |
 |
Structure Based Engineering Of Streptavidin Monomer With A Reduced Biotin Dissociation Rate
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-05-29 Classification: PROTEIN BINDING Ligands: BTN, ZN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-12-05 Classification: Apoptosis/Inhibitor Ligands: 4BP |
 |
Structure Of Base N-Terminal Domain From Acinetobacter Baumannii Bound To 6-(P-Benzyloxy)Phenyl-1-(Pyridin-4-Ylmethyl)-1H-Pyrazolo[3,4-B]Pyridine-4-Carboxylic Acid.
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-10-03 Classification: LIGASE Ligands: H89, CA, MPD, MRD |

