Search Count: 41
 |
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2020-06-24 Classification: BIOSYNTHETIC PROTEIN Ligands: BR, CL |
 |
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-06-24 Classification: BIOSYNTHETIC PROTEIN Ligands: CL, K, BR, EPE |
 |
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2020-06-24 Classification: BIOSYNTHETIC PROTEIN Ligands: QXG |
 |
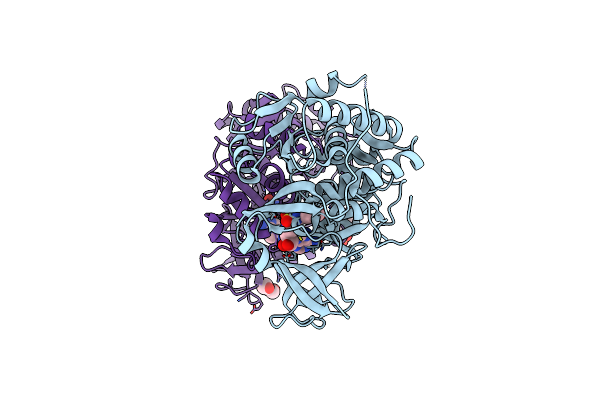
Klebsiella Oxytoca Npsa N-Terminal Subdomain In Complex With 3-Hydroxybenzoyl-Amsn
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2020-06-24 Classification: BIOSYNTHETIC PROTEIN Ligands: R2V, PGE, EPE, PEG, BR, CL |
 |
Klebsiella Oxytoca Npsa N-Terminal Subdomain In Complex With 3-Hydroxyanthranilyl-Amsn
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-06-24 Classification: BIOSYNTHETIC PROTEIN Ligands: QXG, BR, CL, EPE, PEG |
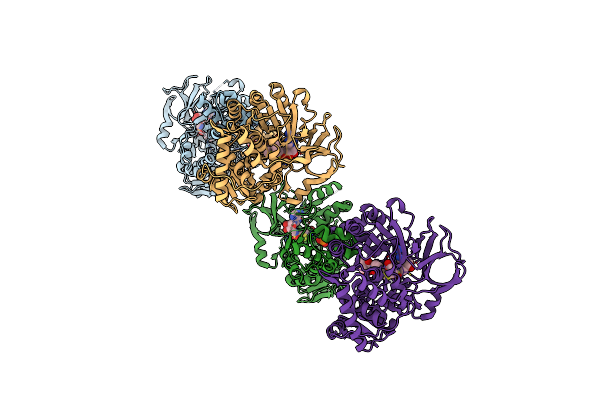 |
Npsa-Thda, An Artificially Fused Adenylation-Pcp Di-Domain Nrps From Klebsiella Oxytoca
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-06-24 Classification: BIOSYNTHETIC PROTEIN Ligands: QXD |
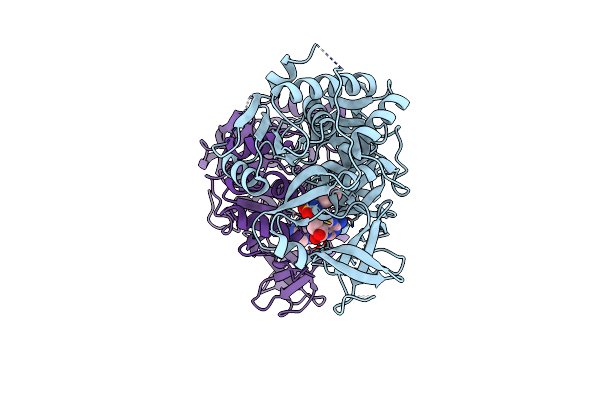 |
Klebsiella Oxytoca Npsa N-Terminal Subdomain In Complex With Anthranilyl-Amsn
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2020-06-24 Classification: BIOSYNTHETIC PROTEIN Ligands: QXP, CL, BR |
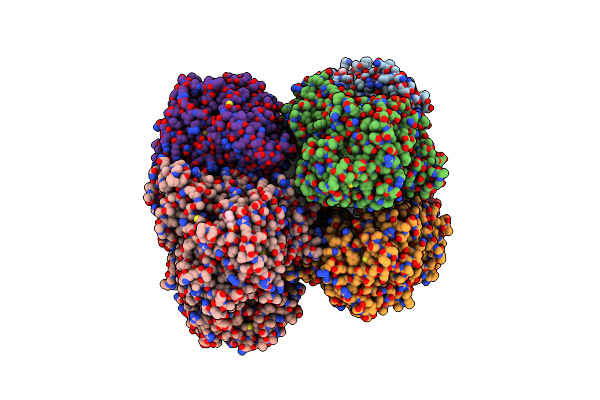 |
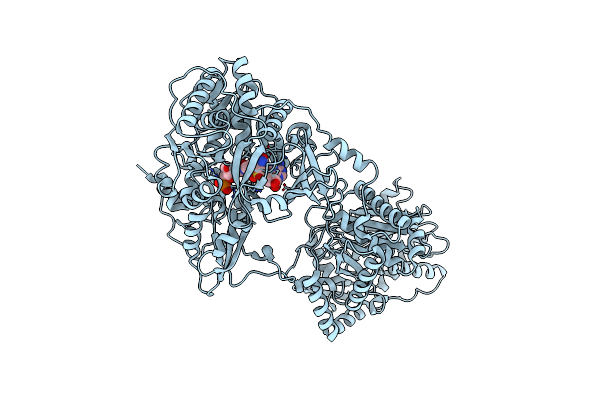
The Structure Of Aerobactin Synthetase Iucc From A Hypervirulent Pathotype Of Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-04-11 Classification: LIGASE Ligands: MES |
 |
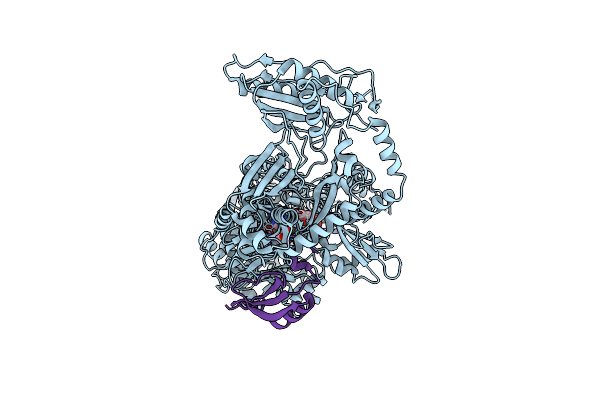
Crystal Structure Of Holo-Entf A Nonribosomal Peptide Synthetase In The Thioester-Forming Conformation
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-09-21 Classification: BIOSYNTHETIC PROTEIN Ligands: 75C |
 |
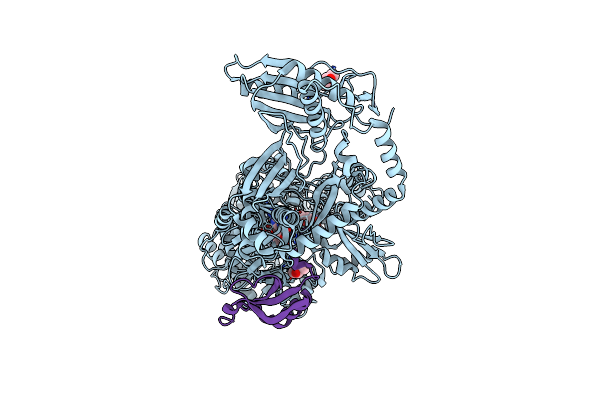
Entf, A Terminal Nonribosomal Peptide Synthetase Module Bound To The Mbth-Like Protein Ybdz
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-09-14 Classification: LIGASE Ligands: 75C, CL |
 |
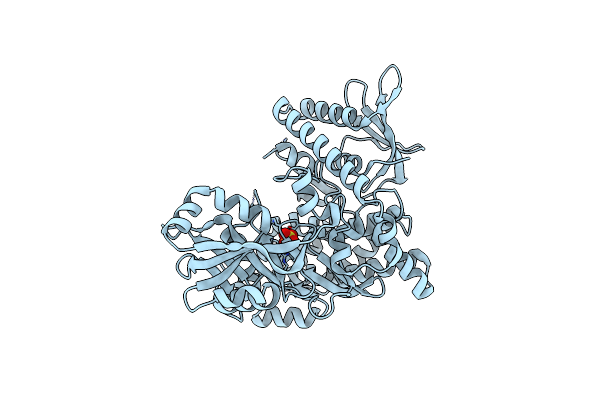
Entf, A Terminal Nonribosomal Peptide Synthetase Module Bound To The Non-Native Mbth-Like Protein Pa2412
Organism: Escherichia coli (strain k12), Pseudomonas aeruginosa (strain atcc 15692 / pao1 / 1c / prs 101 / lmg 12228)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-09-14 Classification: LIGASE Ligands: 75C, BU9 |
 |
The Structure Of Aerobactin Synthetase Iuca From A Hypervirulent Pathotype Of Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-06-15 Classification: LIGASE Ligands: SO4 |
 |
The Structure Of Atp-Bound Aerobactin Synthetase Iuca From A Hypervirulent Pathotype Of Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-06-15 Classification: LIGASE Ligands: ATP, MG |
 |
Structure-Function Studies On Role Of Hydrophobic Clamping Of A Basic Glutamate In Catalysis By Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2016-05-18 Classification: ISOMERASE |
 |
Structure-Function Studies On Role Of Hydrophobic Clamping Of A Basic Glutamate In Catalysis By Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2016-05-18 Classification: ISOMERASE |
 |
Structure-Function Studies On Role Of Hydrophobic Clamping Of A Basic Glutamate In Catalysis By Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-05-18 Classification: ISOMERASE Ligands: K, PGA |
 |
Structure-Function Studies On Role Of Hydrophobic Clamping Of A Basic Glutamate In Catalysis By Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-05-18 Classification: ISOMERASE Ligands: PGA |
 |
Structure-Function Studies On Role Of Hydrophobic Clamping Of A Basic Glutamate In Catalysis By Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-05-18 Classification: ISOMERASE |
 |
Structure-Function Studies On Role Of Hydrophobic Clamping Of A Basic Glutamate In Catalysis By Triosephosphate Isomerase
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2016-05-18 Classification: ISOMERASE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2016-05-11 Classification: TRANSFERASE Ligands: PLP, EDO |

