Search Count: 21
 |
Organism: Pseudomonas phage luz24
Method: SOLUTION NMR Release Date: 2021-05-12 Classification: PEPTIDE BINDING PROTEIN |
 |
Organism: Francisella tularensis subsp. tularensis (strain schu s4 / schu 4), Francisella tularensis
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2020-11-11 Classification: TRANSCRIPTION Ligands: MG, G4P |
 |
Organism: Francisella tularensis subsp. holarctica (strain lvs), Francisella tularensis subsp. holarctica lvs
Method: ELECTRON MICROSCOPY Release Date: 2020-11-11 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Francisella tularensis subsp. holarctica lvs, Francisella tularensis subsp. holarctica (strain lvs)
Method: ELECTRON MICROSCOPY Release Date: 2020-11-11 Classification: TRANSCRIPTION Ligands: MG, ZN |
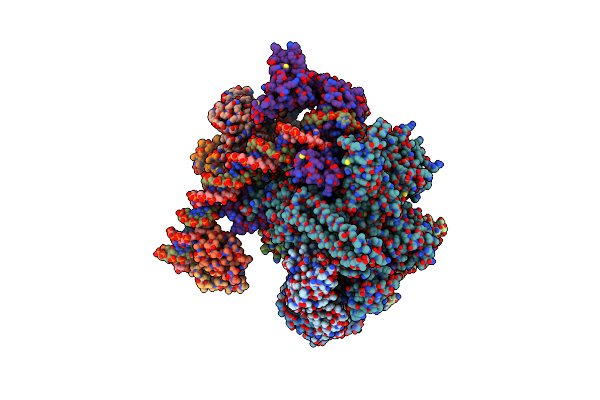 |
Organism: Francisella tularensis subsp. holarctica lvs, Francisella tularensis subsp. holarctica (strain lvs)
Method: ELECTRON MICROSCOPY Release Date: 2020-11-11 Classification: TRANSCRIPTION Ligands: MG, ZN, G4P |
 |
Organism: Escherichia coli, Escherichia coli ta054
Method: ELECTRON MICROSCOPY Release Date: 2020-11-11 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
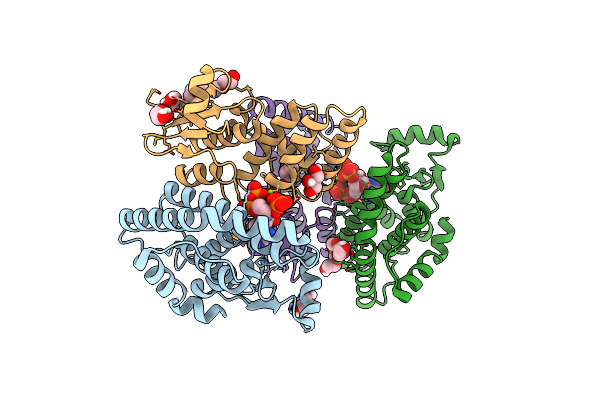
Organism: Francisella tularensis
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-08-23 Classification: TRANSCRIPTION |
 |
Structure Of Francisella Tularensis Heterodimeric Sspa (Mgla-Sspa) In Complex With Ppgpp
Organism: Francisella tularensis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-08-16 Classification: TRANSCRIPTION Ligands: GOL, 1PE, MG, G4P |
 |
Organism: Francisella tularensis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-08-16 Classification: TRANSCRIPTION Ligands: GOL, 1PE, PRO |
 |
Organism: Pseudomonas aeruginosa pao1
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: TRANSCRIPTION REGULATOR |
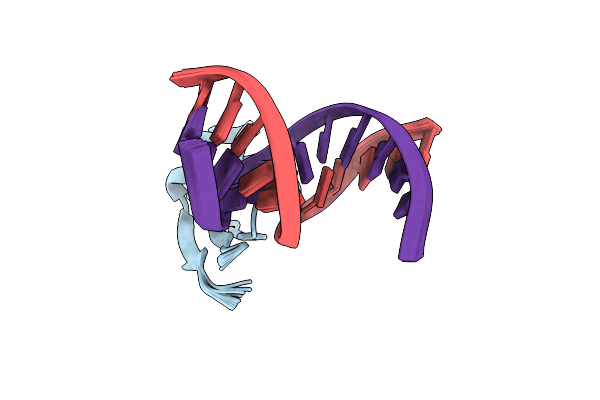 |
Organism: Pseudomonas aeruginosa pao1
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: TRANSCRIPTION REGULATOR/DNA |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2012-03-21 Classification: TSE2-BINDING PROTEIN |
 |
Crystal Structure Of The Tex Protein From Pseudomonas Aeruginosa, Crystal Form I
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2008-04-08 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of The Tex Protein From Pseudomonas Aeruginosa, Crystal Form 2
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-04-08 Classification: RNA BINDING PROTEIN |
 |
Organism: Montipora efflorescens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-04-03 Classification: LUMINESCENT PROTEIN Ligands: IOD |
 |
The 2.0 Angstroms Crystal Structure Of A Pocilloporin At Ph 3.5: The Structural Basis For The Linkage Between Color Transition And Halide Binding
Organism: Montipora efflorescens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-09-05 Classification: LUMINESCENT PROTEIN Ligands: IOD, CL, ACY |
 |
Organism: Pontellina plumata
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-08-15 Classification: LUMINESCENT PROTEIN |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-04-24 Classification: LYASE Ligands: SIE, XLS, SO4 |
 |
Crystal Structure Of S. Pneumoniae Hyaluronate Lyase In Complex With Palmitoyl-Vitamin C
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2004-08-26 Classification: LYASE Ligands: PVC, XYL, SO4 |
 |
Organism: Montipora efflorescens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2003-04-08 Classification: LUMINESCENT PROTEIN Ligands: IOD |

