Search Count: 24
 |
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-01-25 Classification: HYDROLASE |
 |
Organism: Lactobacillus gasseri
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: TAU, K |
 |
Bile Salt Hydrolase A From Lactobacillus Gasseri With Chenodeoxycholate And Taurine Bound
Organism: Lactobacillus gasseri
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: TAU, JN3 |
 |
Organism: Lactobacillus gasseri
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: MG |
 |
Organism: Lactobacillus johnsonii
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-01-25 Classification: HYDROLASE |
 |
Organism: Lactobacillus ingluviei
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: PG4, PEG, PGE, CA |
 |
Organism: Lactobacillus reuteri
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: SO4 |
 |
An Asymmetric Unit Map From Electron Cryo-Microscopy Of Haliotis Diversicolor Molluscan Hemocyanin Isoform 1 (Hdh1)
Organism: Haliotis diversicolor
Method: ELECTRON MICROSCOPY Release Date: 2013-04-17 Classification: OXYGEN TRANSPORT |
 |
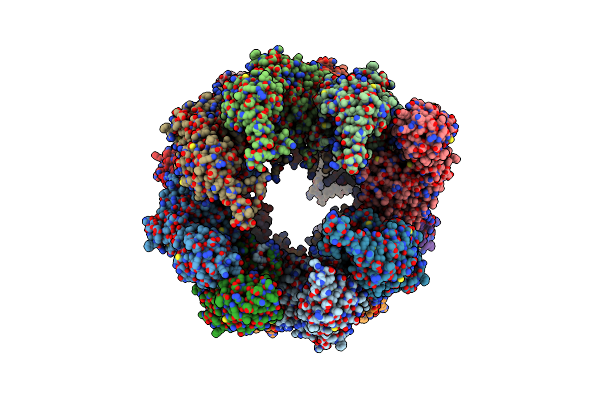
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Resolution:10.50 Å Release Date: 2012-02-15 Classification: CHAPERONE |
 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Resolution:10.70 Å Release Date: 2012-02-15 Classification: CHAPERONE |
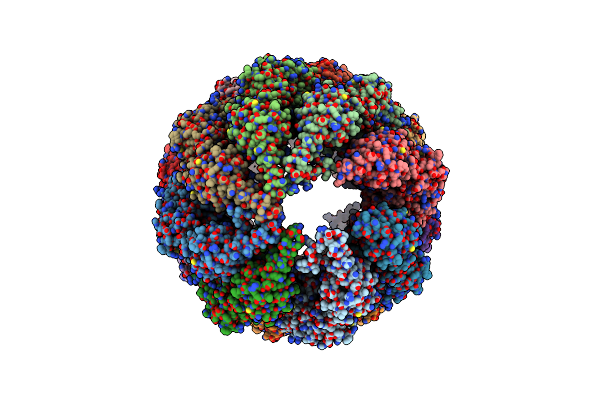 |
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Resolution:13.90 Å Release Date: 2012-02-15 Classification: CHAPERONE |
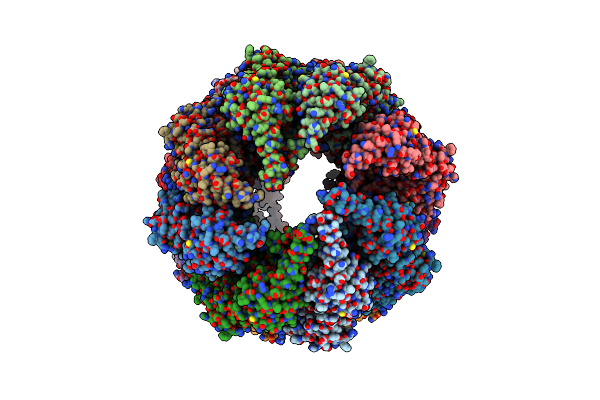 |
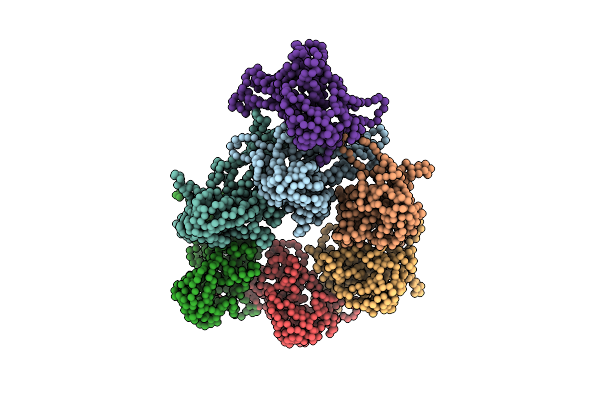
Organism: Bos taurus
Method: ELECTRON MICROSCOPY Resolution:11.30 Å Release Date: 2012-02-15 Classification: CHAPERONE |
 |
Organism: Enterobacteria phage p22
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2011-02-02 Classification: VIRUS |
 |
Organism: Enterobacteria phage p22
Method: ELECTRON MICROSCOPY Resolution:4.00 Å Release Date: 2011-02-02 Classification: VIRUS |
 |
Organism: Prochlorococcus phage p-ssp7
Method: ELECTRON MICROSCOPY Resolution:4.60 Å Release Date: 2010-06-16 Classification: VIRUS |
 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2010-03-16 Classification: CHAPERONE |
 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2010-02-02 Classification: CHAPERONE |
 |
Scorpion Hemocyanin Resting State Pseudo Atomic Model Built Based On Cryo-Em Density Map
Organism: Androctonus australis
Method: ELECTRON MICROSCOPY Release Date: 2009-06-02 Classification: OXYGEN BINDING |
 |
Scorpion Hemocyanin Activated State Pseudo Atomic Model Built Based On Cryo-Em Density Map
Organism: Androctonus australis
Method: ELECTRON MICROSCOPY Release Date: 2009-06-02 Classification: OXYGEN BINDING |
 |
Actin Filament Model In The Extended Form Of Acromsomal Bundle In The Limulus Sperm
Organism: Limulus polyphemus
Method: ELECTRON MICROSCOPY Release Date: 2008-11-18 Classification: CONTRACTILE PROTEIN/STRUCTURAL PROTEIN |

