Search Count: 14
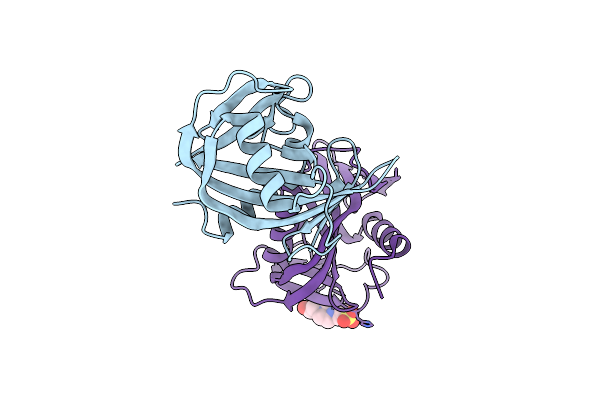 |
Crystal Structure Of The Bacillus Subtilis Pyrophosphohydrolase Bsrpph (E68A Mutant)
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-05-08 Classification: HYDROLASE Ligands: EPE |
 |
Crystal Structure Of The Bacillus Subtilis Pyrophosphohydrolase Bsrpph Bound To A Non-Hydrolysable Triphosphorylated Dinucleotide Rna (Pcp-Pgpg) - First Guanosine Residue In Guanosine Binding Pocket
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-05-08 Classification: HYDROLASE/RNA Ligands: EPE |
 |
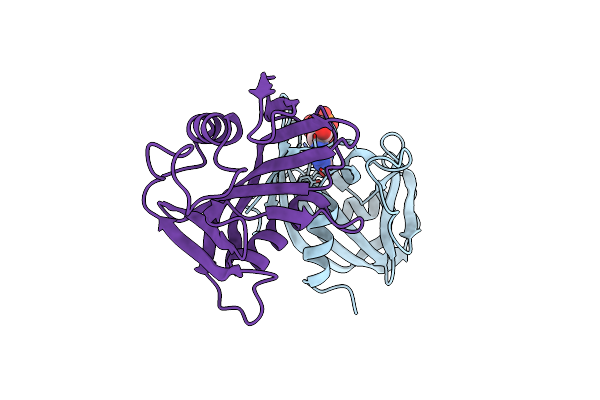
Crystal Structure Of The Bacillus Subtilis Pyrophosphohydrolase Bsrpph Bound To A Non-Hydrolysable Triphosphorylated Dinucleotide Rna (Pcp-Pgpg) - Second Guanosine Residue In Guanosine Binding Pocket
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-05-08 Classification: HYDROLASE/RNA Ligands: MG, EPE |
 |
Crystal Structure Of The Bacillus Subtilis Pyrophosphohydrolase Bsrpph (E68A Mutant) Bound To Gtp
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2013-05-01 Classification: HYDROLASE Ligands: GTP |
 |
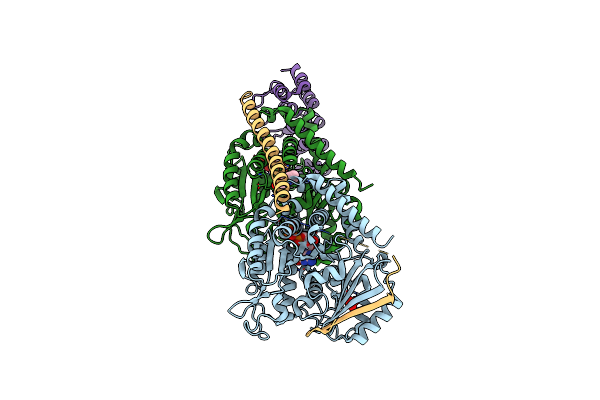
Organism: Artificial gene, Ovis aries
Method: X-RAY DIFFRACTION Resolution:4.17 Å Release Date: 2012-07-18 Classification: CELL CYCLE Ligands: GTP, MG, GDP |
 |
Organism: Artificial gene, Ovis aries
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2012-07-18 Classification: CELL CYCLE Ligands: GTP, MG, SO4, GDP, MES |
 |
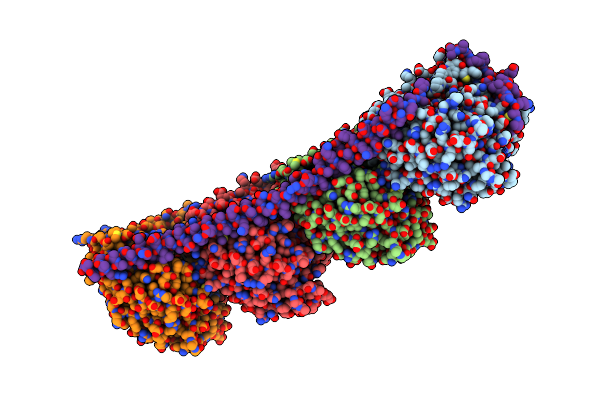
Molecular Basis For The Recognition And Cleavage Of Rna (Uuccgu) By The Bifunctional 5'-3' Exo/Endoribonuclease Rnase J
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2011-10-19 Classification: HYDROLASE/RNA Ligands: ZN |
 |
Molecular Basis For The Recognition And Cleavage Of Rna (Cugg) By The Bifunctional 5'-3' Exo/Endoribonuclease Rnase J
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-10-19 Classification: HYDROLASE/RNA Ligands: ZN, GOL |
 |
Organism: Rattus norvegicus, Ovis aries
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2010-07-28 Classification: CELL CYCLE Ligands: GTP, MG, GDP, G2N |
 |
Organism: Rattus norvegicus, Ovis aries
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2010-07-28 Classification: CELL CYCLE Ligands: GTP, MG, GDP, K2N |
 |
Organism: Rattus norvegicus, Ovis aries
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2009-09-01 Classification: CELL CYCLE Ligands: GTP, MG, GDP |
 |
Organism: Rattus norvegicus, Ovis aries
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2009-09-01 Classification: CELL CYCLE Ligands: GTP, MG, GDP, E70 |
 |
Organism: Rattus norvegicus, Ovis aries
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2009-09-01 Classification: CELL CYCLE Ligands: GTP, MG, GDP, N16 |
 |
Organism: Rattus norvegicus, Ovis aries
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2009-09-01 Classification: CELL CYCLE Ligands: GTP, MG, T13, GDP |

